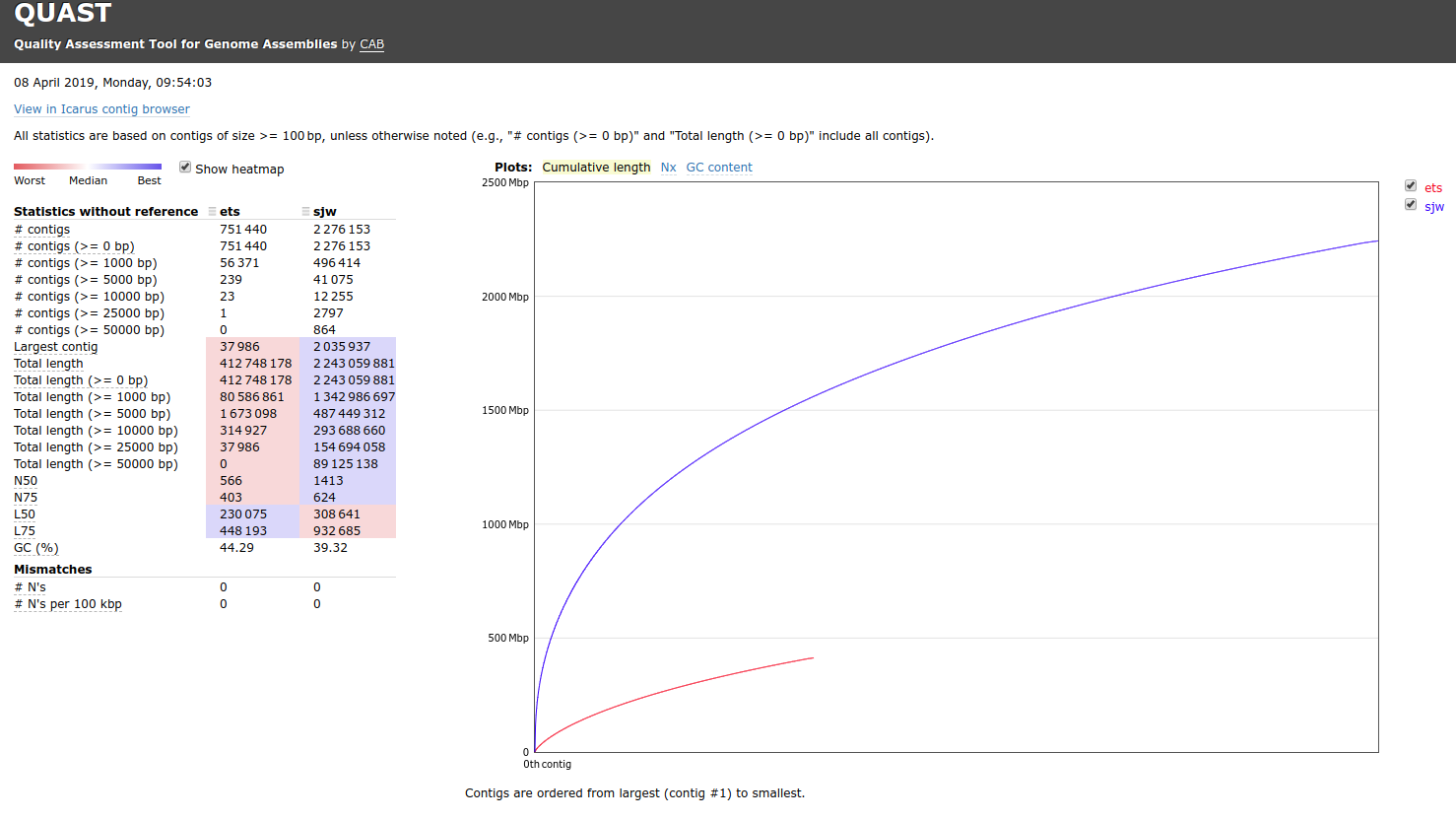
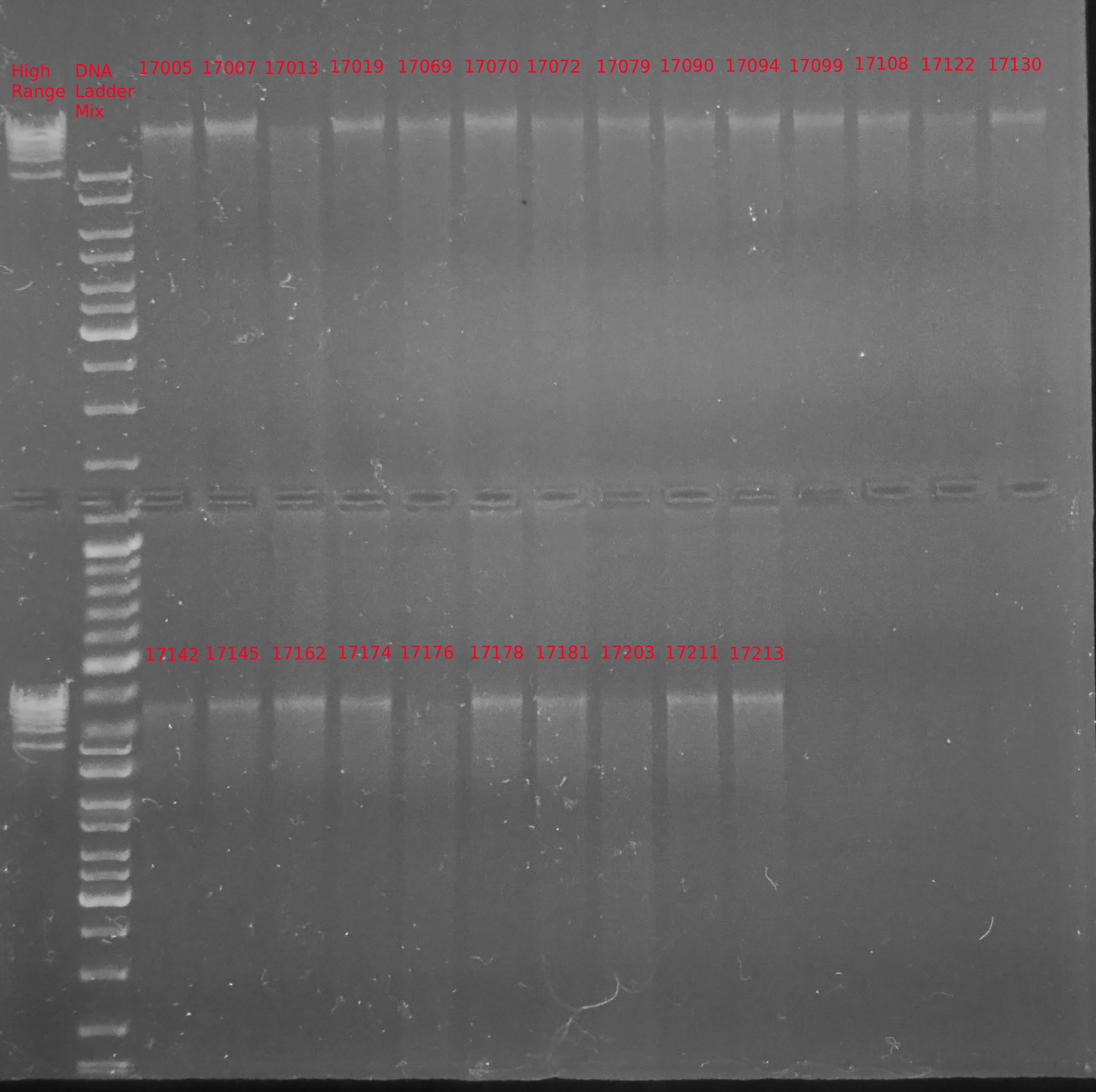
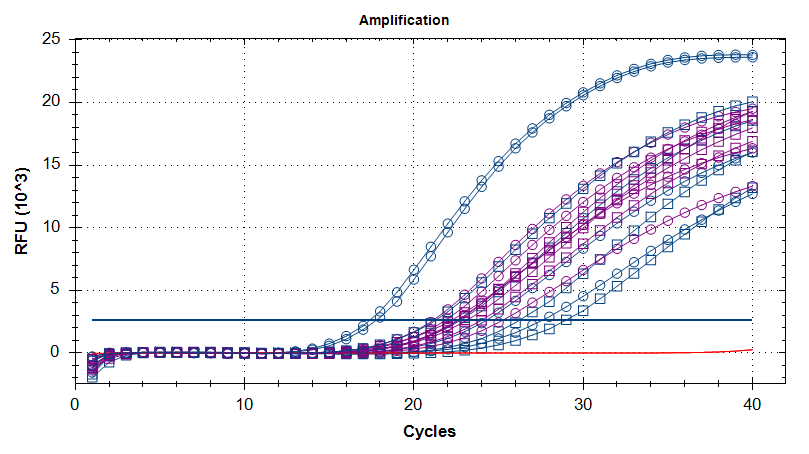
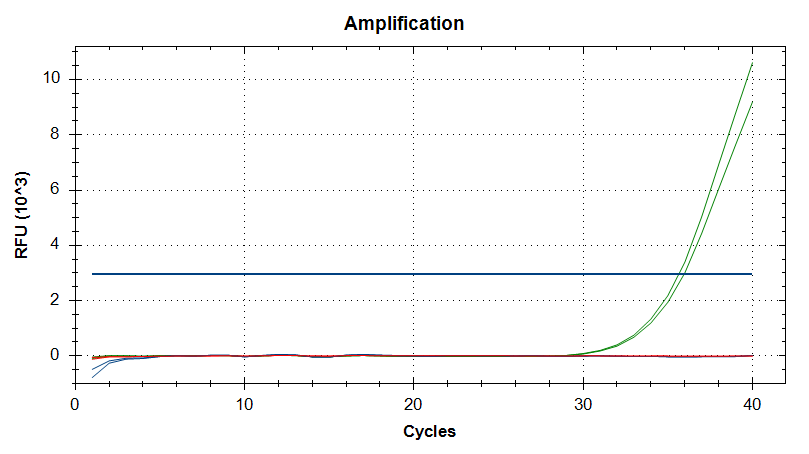
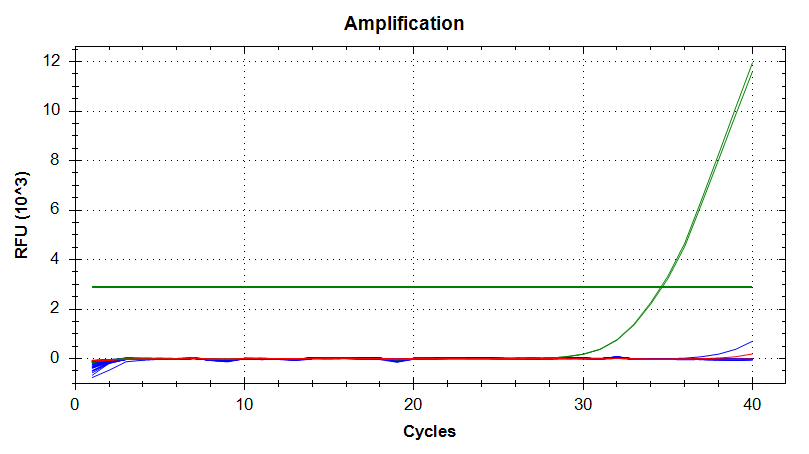
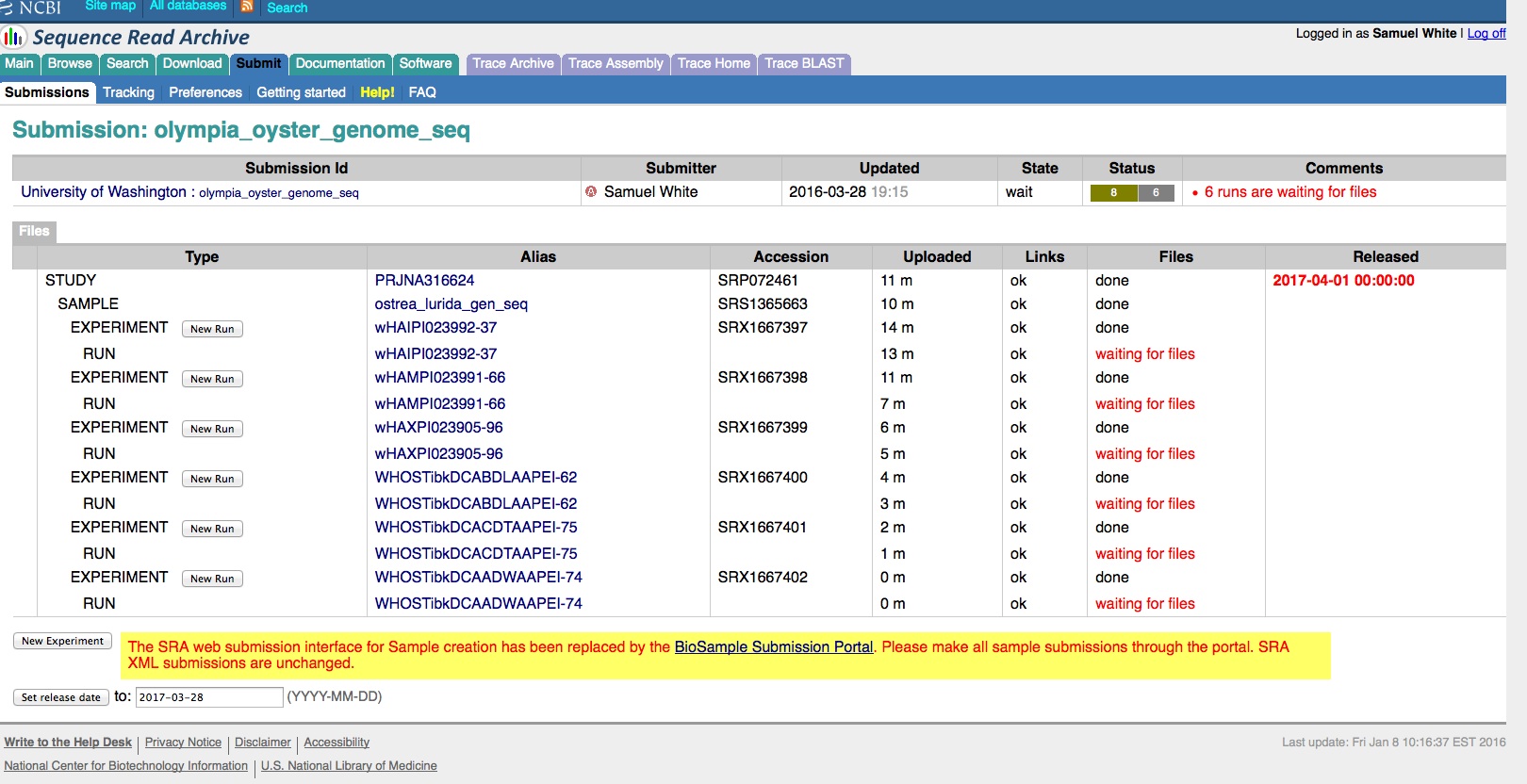
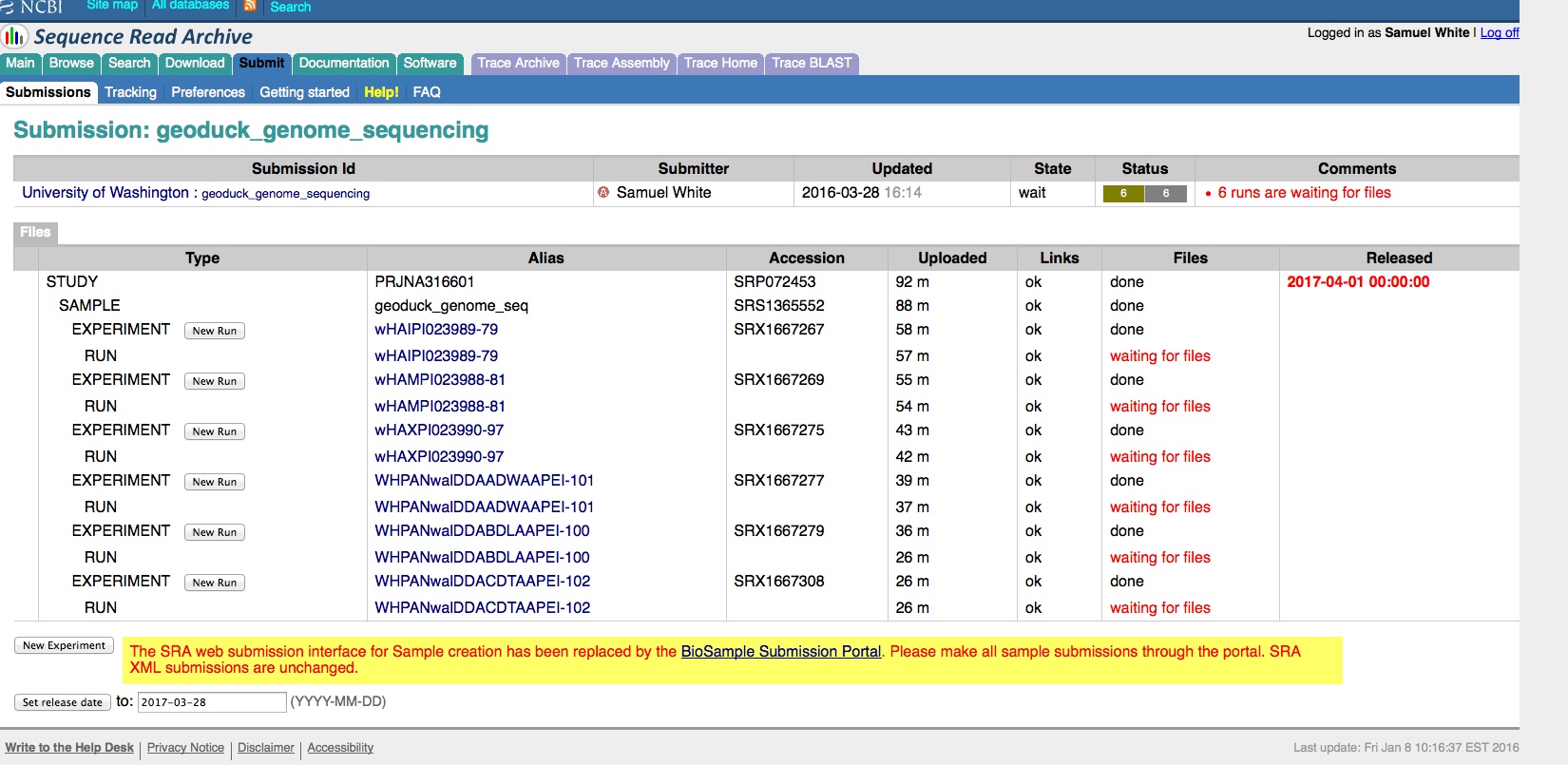
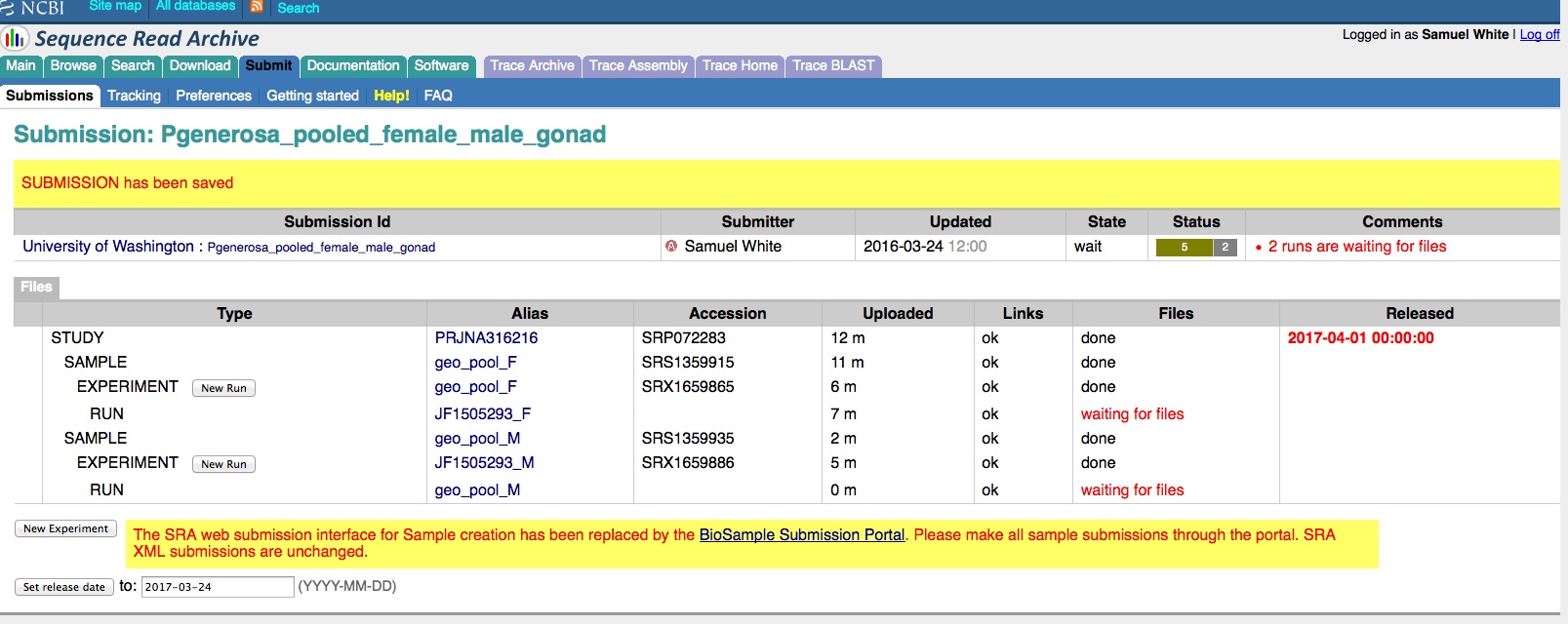
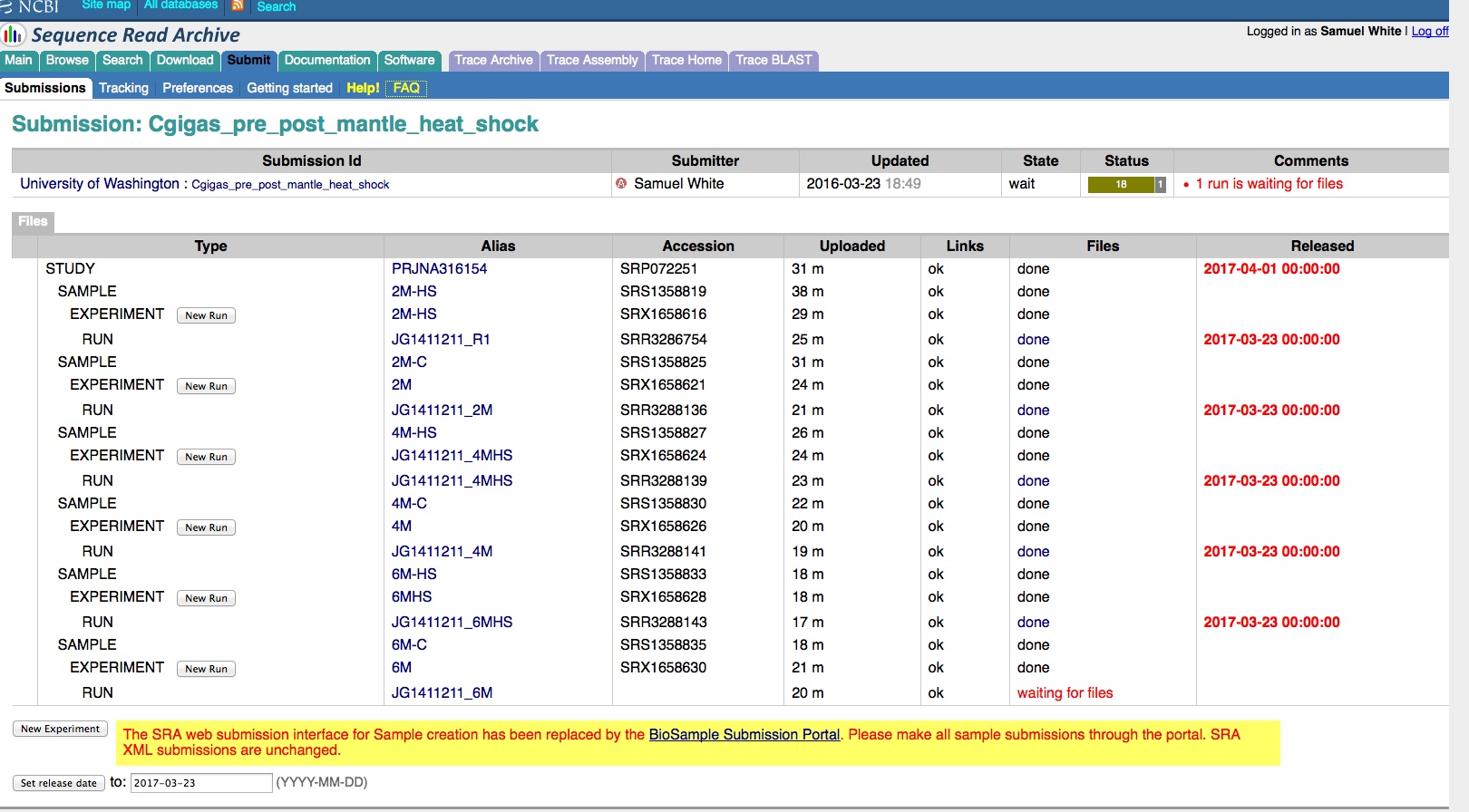
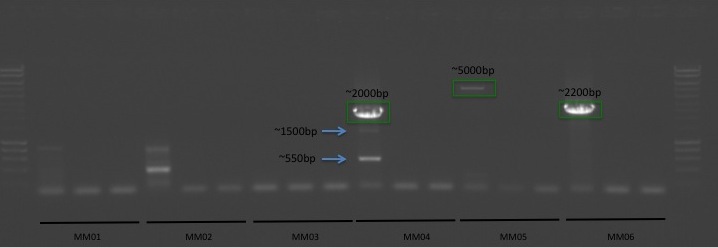
Transcriptome Annotation - M.magister De Novo Transcriptome Assembly Using Trinotate on Mox
mox
2023
Metacarcinus magister
Dungenss crab
Trinotate
transcriptome
annotation
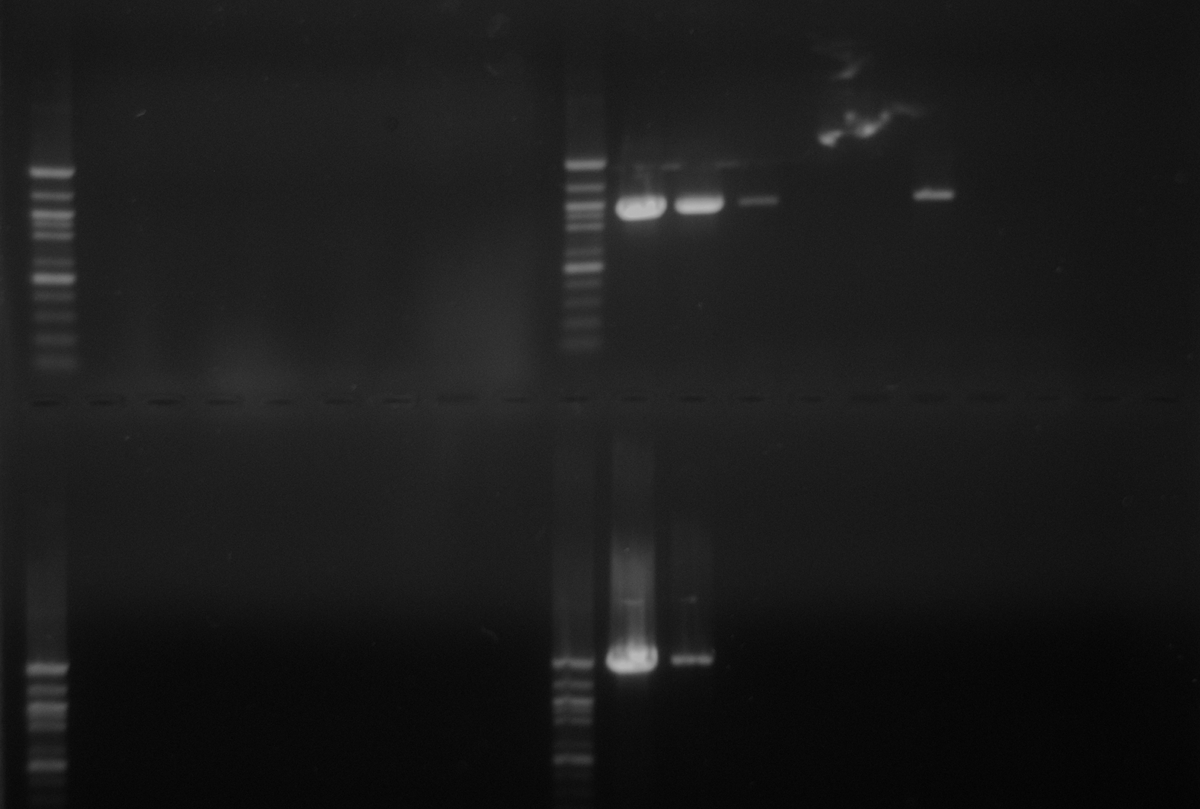
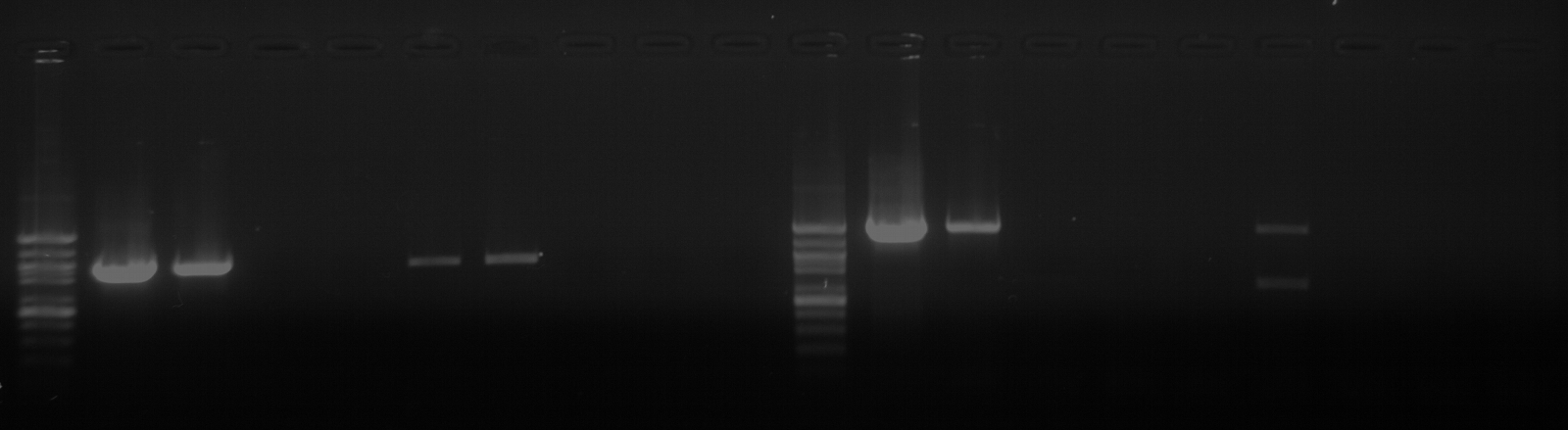
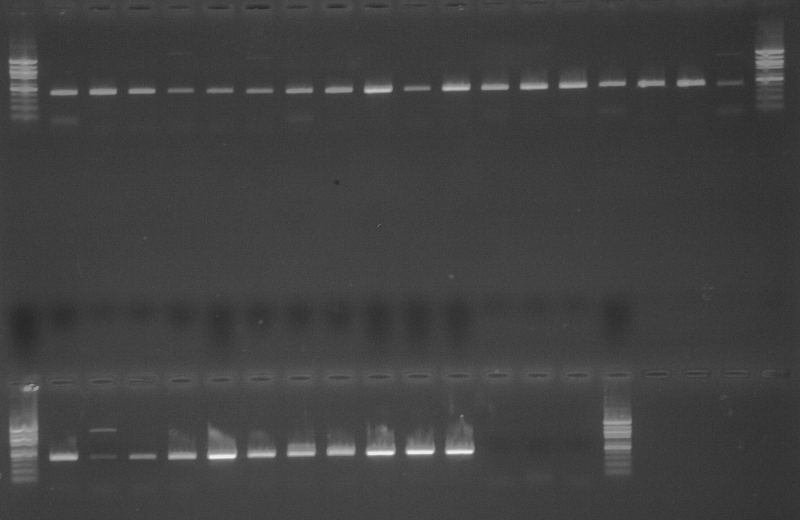
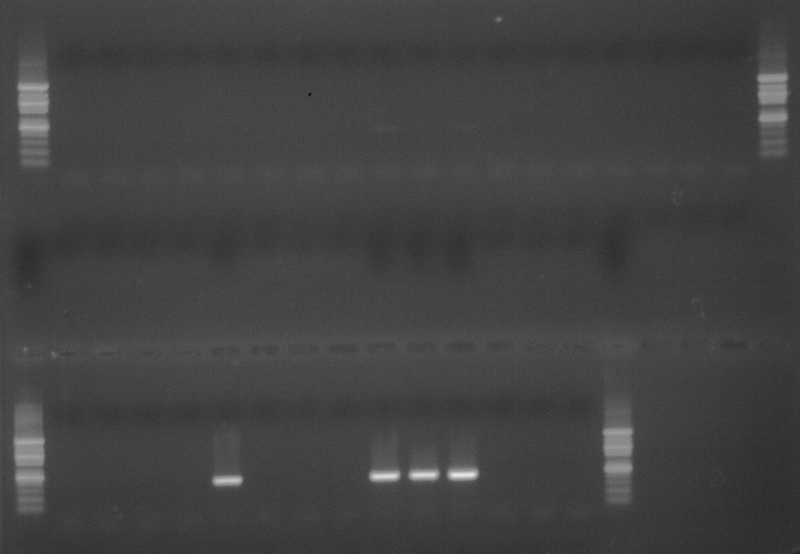
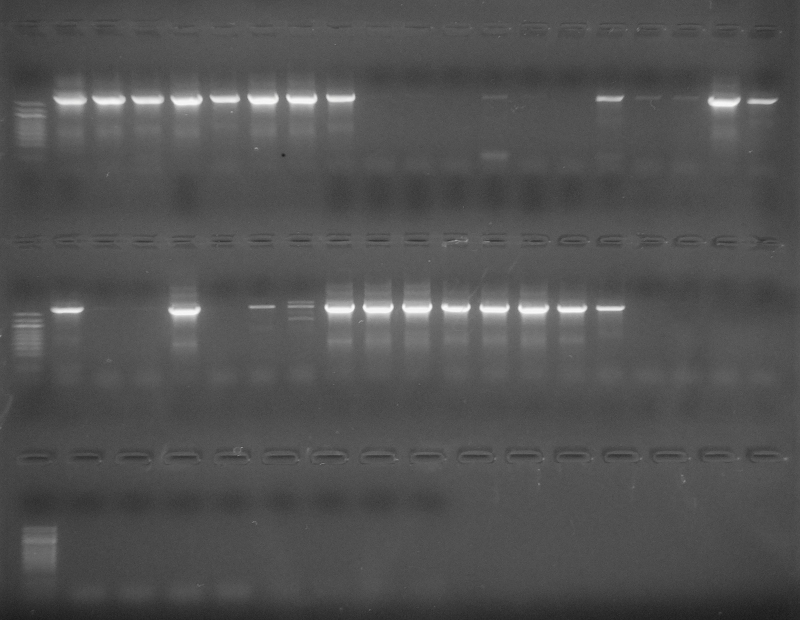
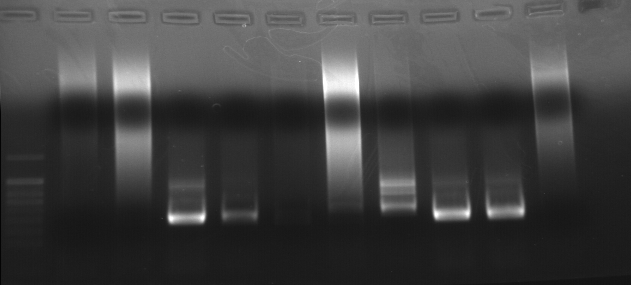
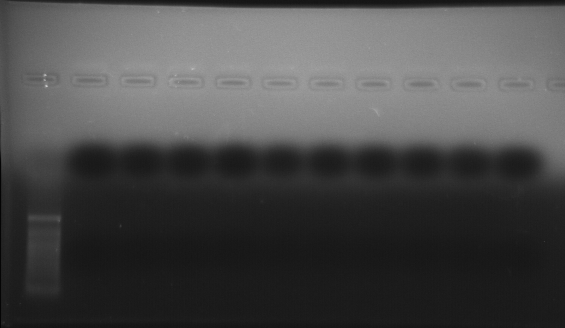
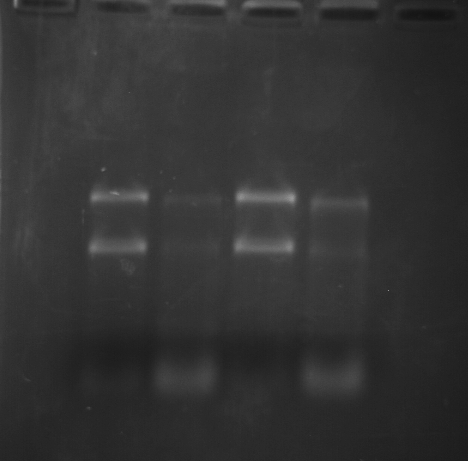
As part of transcriptome annotation (GitHub issue) for DuMOAR (GitHub repo), I’ve already run
TransDecoder ( run on 20231107), as well as DIAMOND BLASTx (run earlier today). The final part of getting the transcriptome annotated was…Transcriptome Annotation - M.magister De Novo Transcriptome Assembly for DuMOAR Project Using DIAMOND BLASTx on Mox
mox
DIAMOND
BLASTx
transcriptome
2023
DuMOAR
Dungeness crab
Metacarcinus magister
As part of the process for using
Trinotate for transcriptome annotation (GitHub Issue), I previously ran TransDecoder on 20231107.…Transcript Identification and Alignments - C.virginica RNAseq with NCBI Genome GCF_002022765.2 Using Hisat2 and Stringtie on Mox Again
2023
Miscellaneous
In the process of generating expression matrices for CEABIGR (GitHub repo), and in turn distance matrices for these samples, I realized a couple of things:
Reverse Transcription - C.gigas RNA from Noah’s Heat-Mechanical Stress Project
2023
Miscellaneous
After isolating ctenidia RNA on 20230712 and then quantifying the RNA earlier today, I needed to perform reverse transcription to have cDNA for subsequent qPCRs. Reverse transcription was performed using oligo dT primers using M-MLV RT (Promega), per the…
ORF Identification - L.staminea De Novo Transcriptome Assembly v1.0 Using Transdecoder on Mox
2023
Miscellaneous
After performing a de novo transcriptome assembly with L.staminea RNA-seq data, the
Trinity assembly stats were quite a bit more “exaggerated” than normally expected. In an attempt to get a better sense of which contigs might be…
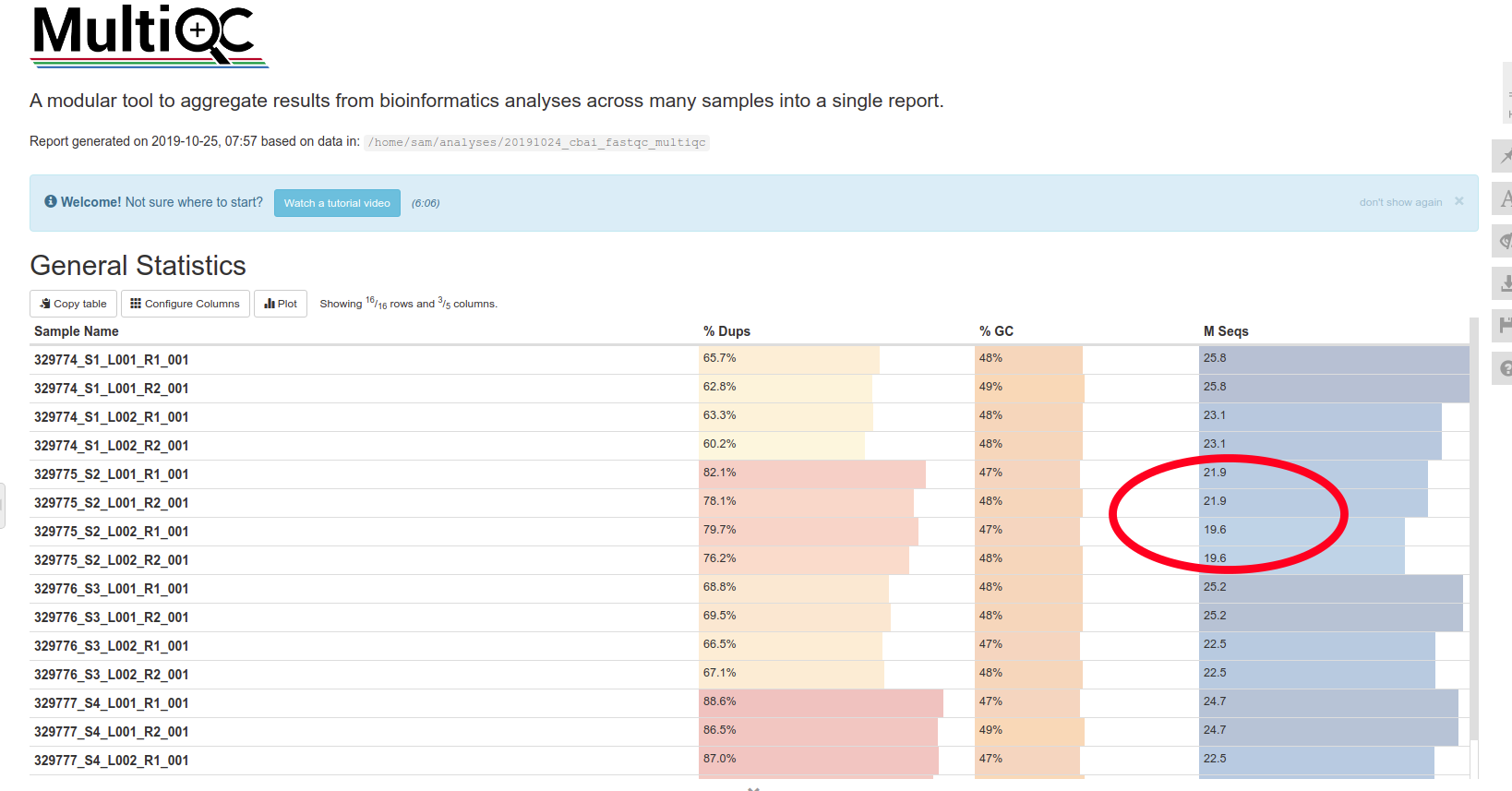
FastQ QC and Trimming - E5 Coral RNA-seq Data for A.pulchra P.evermanni and P.meandrina Using FastQC fastp and MultiQC on Mox
2023
E5
After downloading and then reorganizing the E5 coral RNA-seq data from Azenta project 30-789513166, I ran FastQC for initial quality checks, followed by trimming with
fastp, and then final QC with FastQC/MultiQC. This was performed on all three species in the data…
Data Management - E5 Coral RNA-seq and sRNA-seq Reorganizing and Renaming
2023
Data Management
Downloaded the E5 coral sRNA-seq data from Azenta project 30-852430235 on 20230515 and the E5 coral RNA-seq data from Azenta project 30-789513166 on 20230516. The data required some reorganization, as the project included data from three…
Containers - Apptainer Explorations
2023
Miscellaneous
At some point, our HPC nodes on Mox will be retired. When that happens, we’ll likely purchase new nodes on the newest UW cluster, Klone. Additionally, the
coenv nodes are no longer available on Mox. One was decommissioned and one was “migrated” to Klone. The primary issue at hand is that the base…
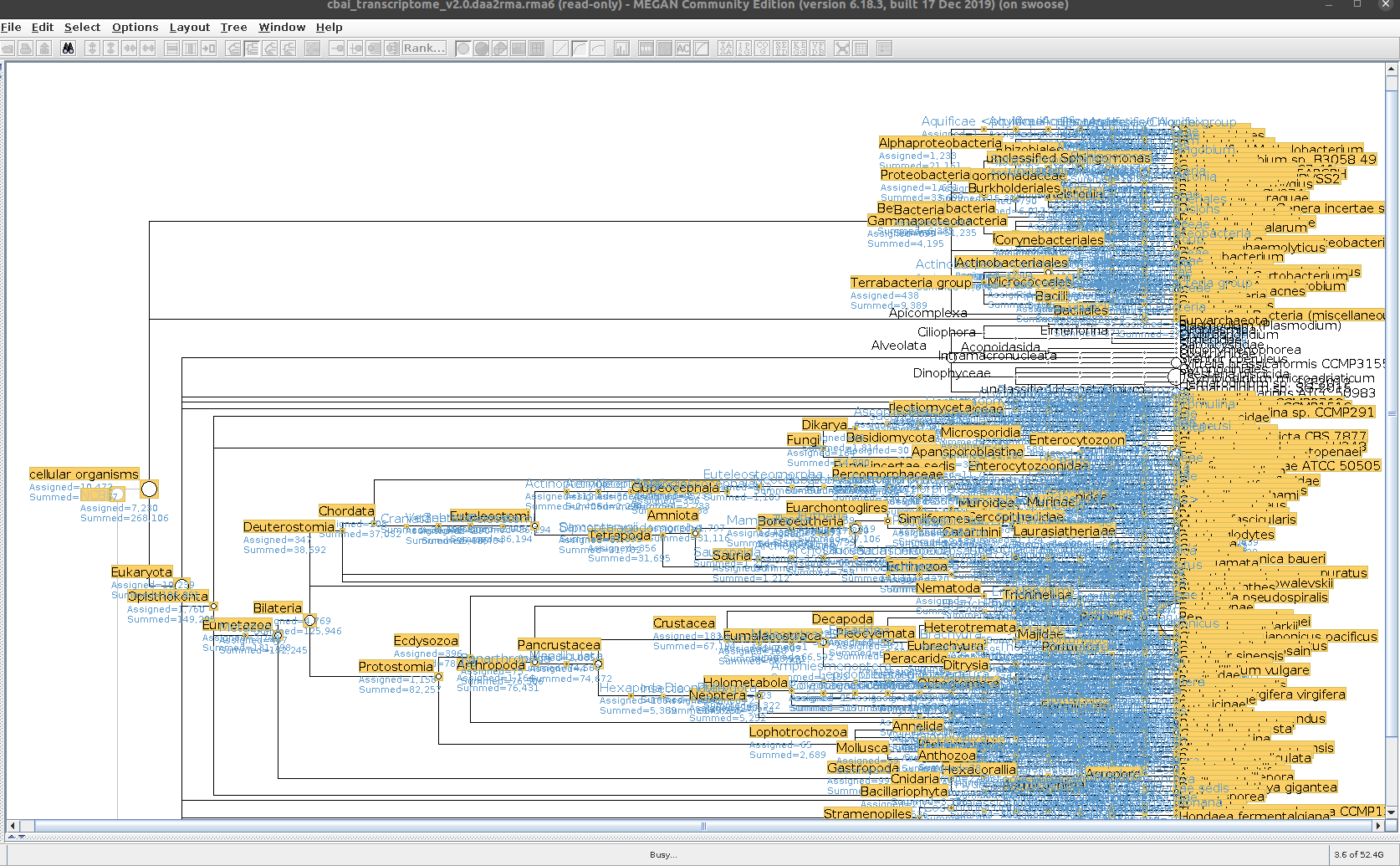
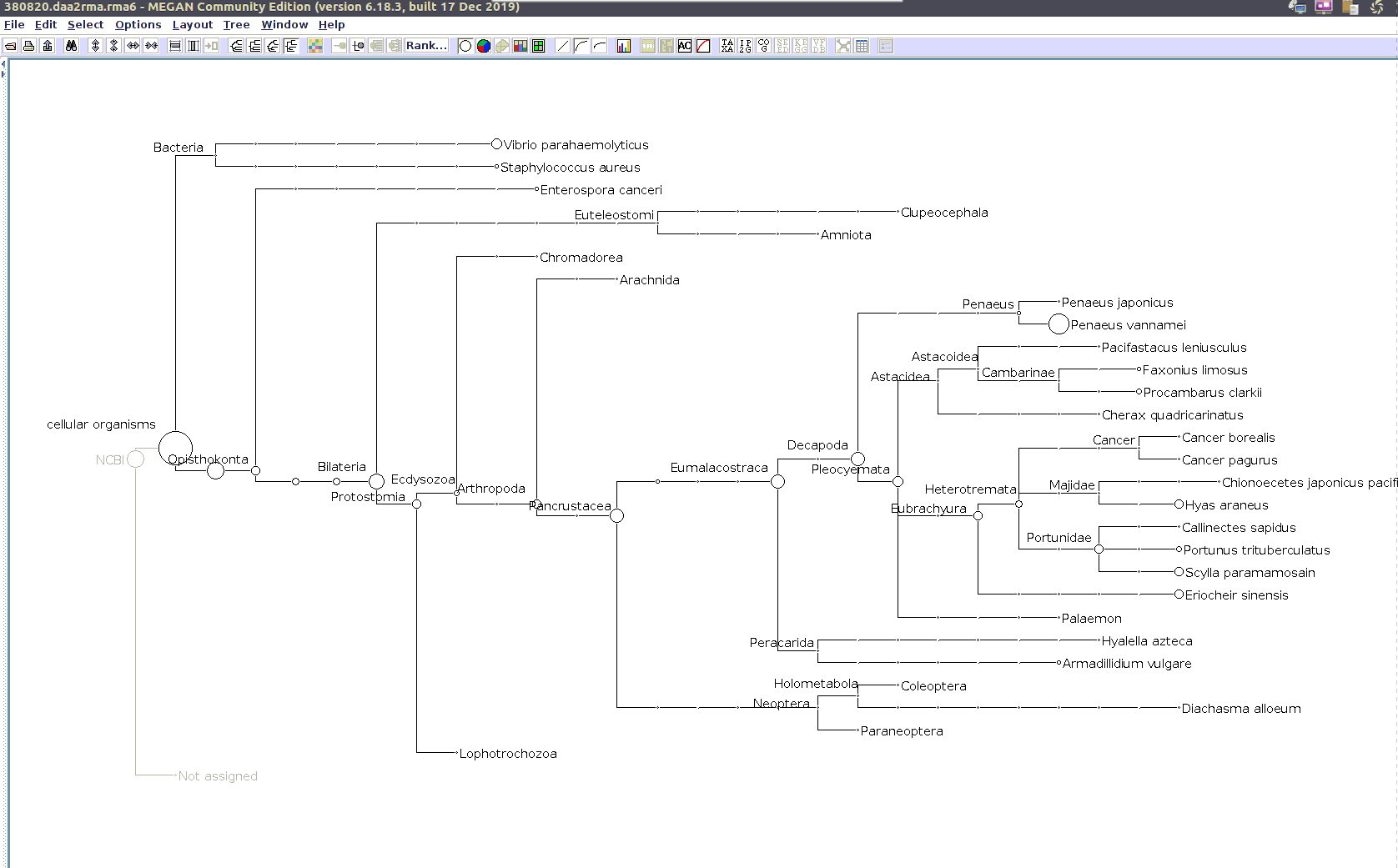
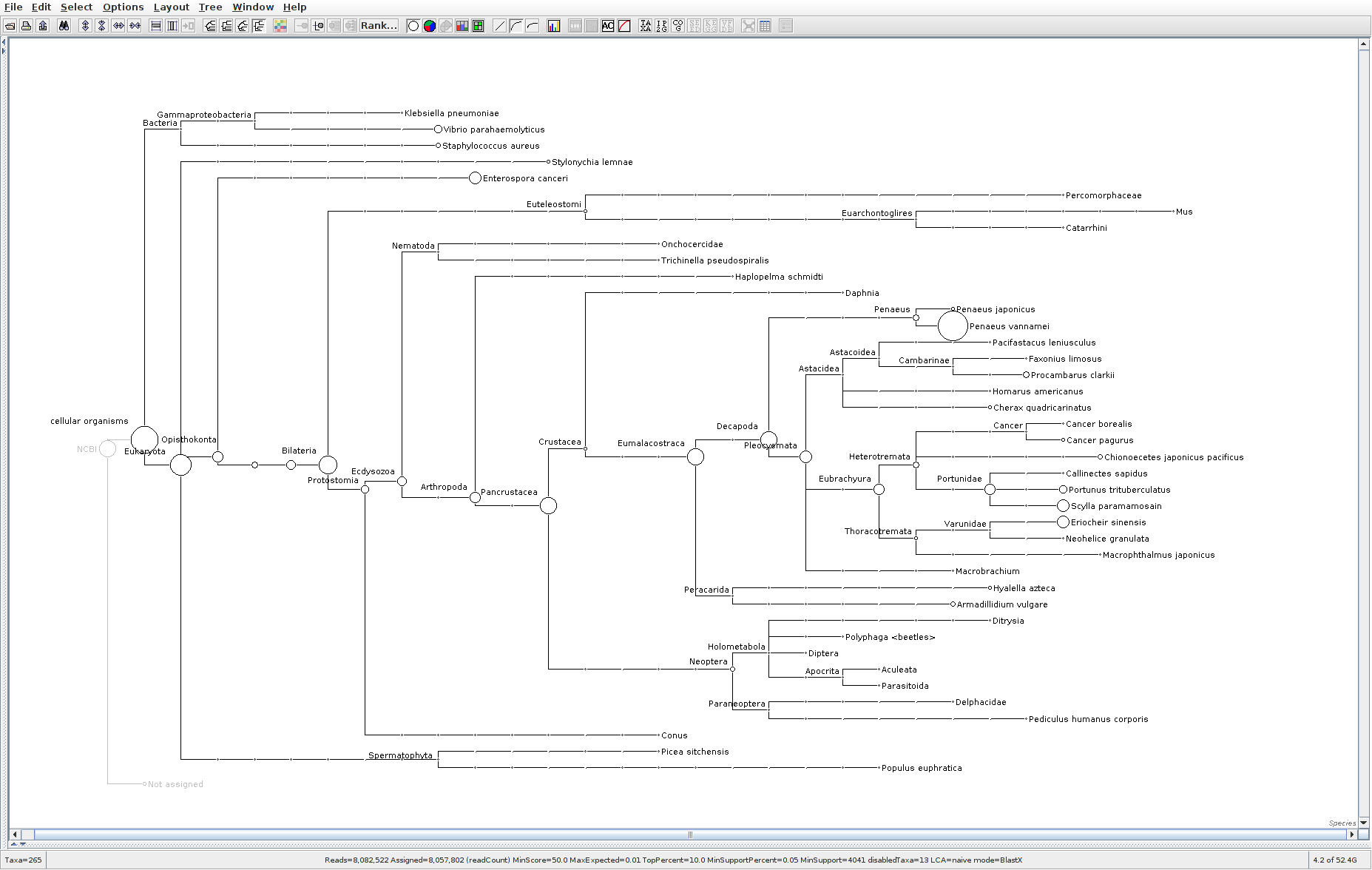
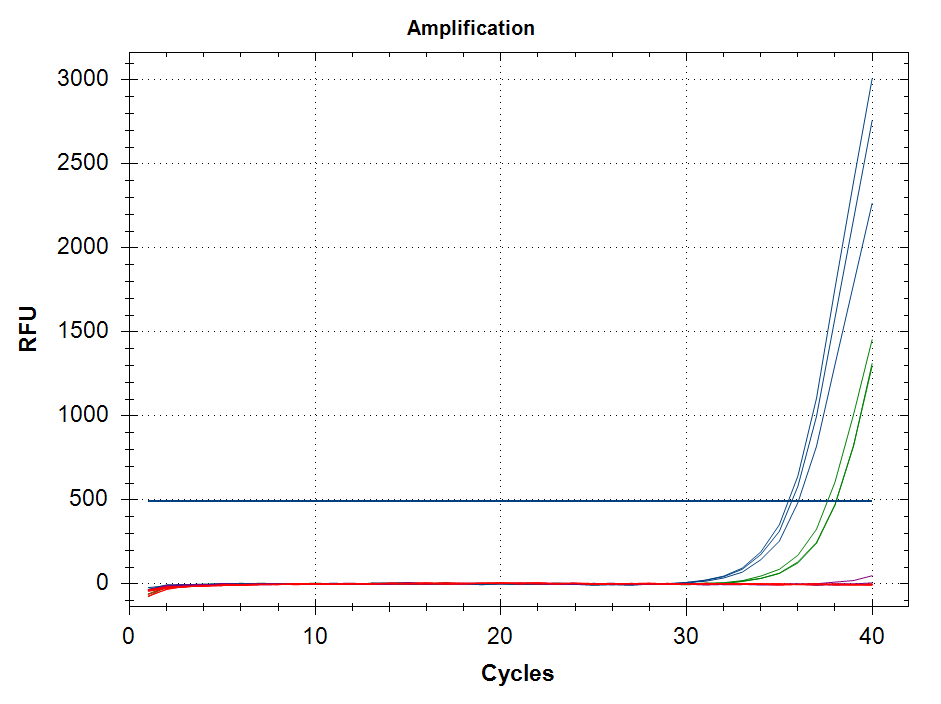
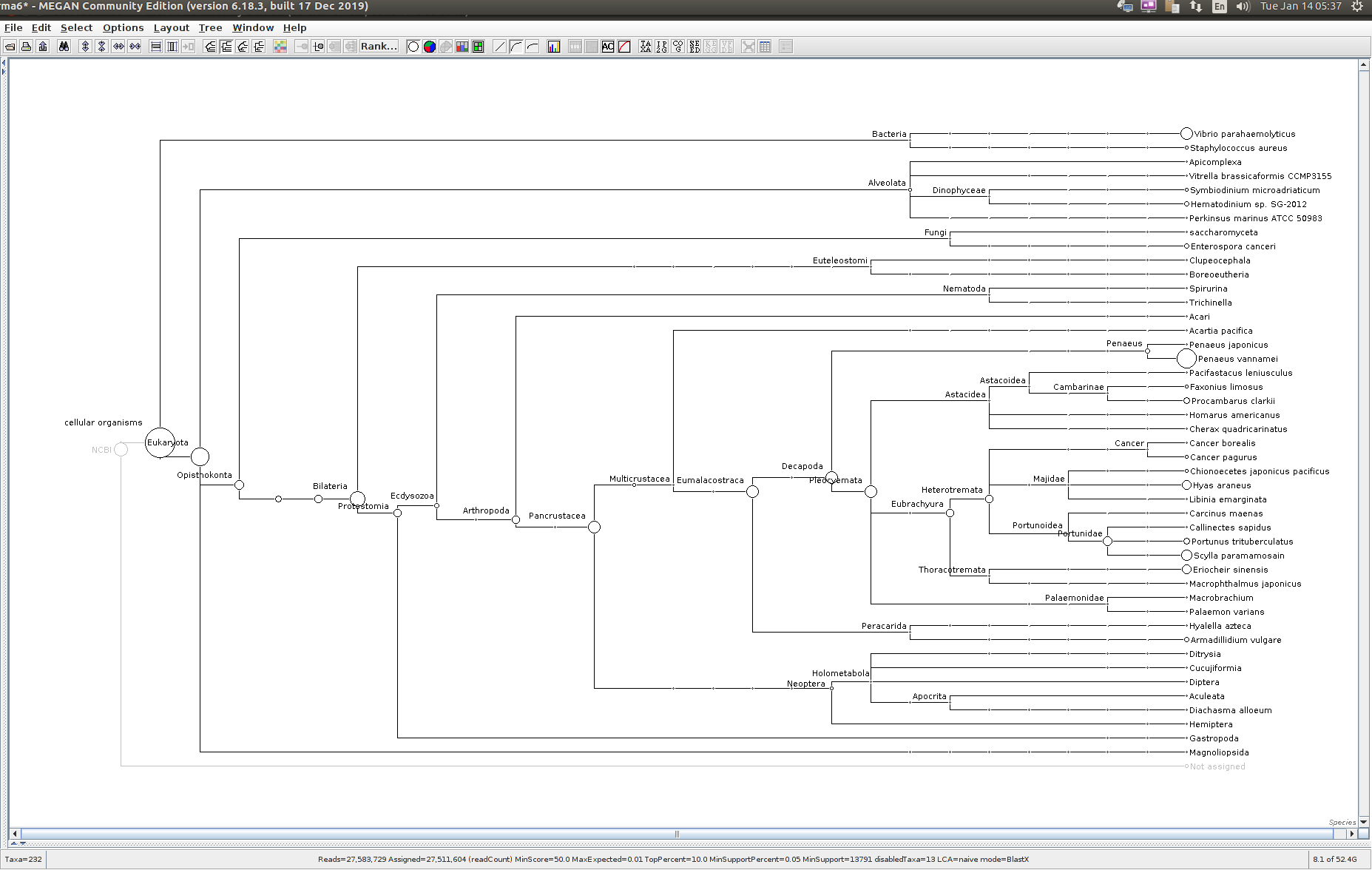
Sequencing Read Taxonomic Classification - M.magister RNA-seq Using DIAMOND BLASTx and MEGAN6 daa-meganizer on Mox
2023
Miscellaneous
Running DIAMOND BLASTx, followed by MEGAN6
daa-meganizer for taxonomic classification of NOAA M.magister trimmed RNA-seq reads (provided by Giles Goetz on 20230301). This is primarily just for…
FastQ Trimming and QC - P.verrucosa RNA-seq Data from Danielle Becker in Hollie Putnam Lab Using fastp FastQC and MultiQC on Mox
2023
E5
After receiving the P.verrucosa RNA-seq data from Danielle Becker (Hollie Putnam’s Lab, Univ. of Rhode Island), I noticed that the trimmed reads didn’t appear to actually be trimmed. There was still adapter contamination…
Data Received - P.verrucosa RNA-seq and WGBS Full Data from Danielle Becker
2023
E5
Data Received
Worked with Danielle Becker, as part of the Coral E5 project, to transfer data related to her repo, from her HPC (Univ. of Rhode Island; Andromeda) to ours (Univ. of Washington; Mox) in order to eventually transfer to Gannet so…
Data Wrangling - P.acuta Genome GFF to GTF Conversion Using gffread
2023
Miscellaneous
As part of getting these three coral species genome files (GitHub Issue) added to our Lab Handbook Genomic Resources page, I will index the P.acuta genome file using
HISAT2, but need a GTF file to also identify exon/intro splice sites. Since a GTF file is not available, but a GFF file is, I needed to convert the GFF to GTF.…Data Wrangling - C.virginica NCBI GCF_002022765.2 GFF to Gene and Pseudogene Combined BED File
2022
Miscellaneous
Working on the CEABIGR project, I was preparing to make a gene expression file to use in CIRCOS (GitHub Issue) when I realized that the Ballgown gene expression file (CSV; GitHub) had more…
BSseq SNP Analysis - Nextflow EpiDiverse SNP Pipeline for C.virginica CEABIGR BSseq data
2022
Miscellaneous
Steven asked that I identify SNPs from our C.virginica CEABIGR BSseq data (GitHub Issue). So, I ran sorted, deduplicated Bismark BAMs that Steven generated through the EpiDiverse/snp Nextflow…
RNAseq Alignments - P.generosa Alignments and Alternative Transcript Identification Using Hisat2 and StringTie on Mox
2022
Miscellaneous
As part of identifying long non-coding RNA (lncRNA) in Pacific geoduck(GitHub Issue), one of the first things that I wanted to do was to gather all of our geoduck RNAseq data and align it to our geoduck genome. In addition to the alignments…
FastQ Trimming - Geoduck RNAseq Data Using fastp on Mox
2022
Miscellaneous
Per this GitHub Issue, Steven asked me to identify long non-coding RNA (lncRNA) in geoduck. The first step is to aggregate all of our Panopea generosa (Pacific geoduck) RNAseq data and get it all trimmed. After that, align it to the genome…
FastQ Trimming and QC - C.virginica Larval BS-seq Data from Lotterhos Lab and Part of CEABIGR Project Using fastp on Mox
2022
Miscellaneous
We had some old Crassostrea virginica (Eastern oyster) larval/zygote BS-seq data from the Lotterhos Lab that’s part of the CEABiGR Workshop/Project (GitHub Repo) and…
Splice Site Identification - S.namaycush Liver Parasitized and Non-Parasitized SRA RNAseq Using Hisat2-Stingtie with Genome GCF_016432855.1
2022
Miscellaneous
After previously downloading/trimming/QCing S.namaycush SRA liver RNAseq data on 20220706, Steven asked that I run through Hisat2 for splice site identification…
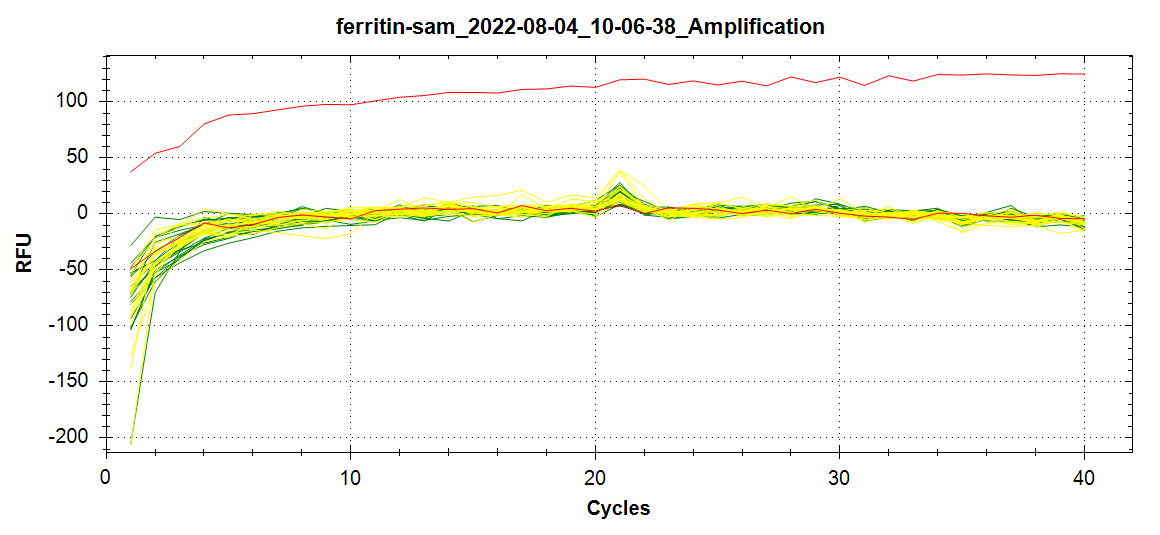
qPCR - Repeat of Mussel Gill Heat Stress cDNA with Ferritin Primers
2022
Miscellaneous
My previous qPCR on these cDNA using ferritin primers (SRIDs: 1808, 1809) resulted in no amplification. This was a bit surprising and makes me suspect that I screwed up somewhere (not adding primer(s)??), so I decided to…
Differential Gene Expression - P.generosa DGE Between Tissues Using Nextlow NF-Core RNAseq Pipeline on Mox
2022
Miscellaneous
Steven asked that I obtain relative expression values for various geoduck tissues (GitHub Issue). So, I decided to use this as an opportunity to try to use a Nextflow pipeline. There’s an RNAseq…
Data Analysis - C.virginica RNAseq Zymo ZR4059 Analyzed by ZymoResearch
2022
Miscellaneous
After realizing that the Crassostrea virginica (Eastern oyster) RNAseq data had relatively low alignment rates (see this notebook entry from 20220224 for a bit more background), I contacted ZymoResearch to see if they had any insight on what might be happening. I suspected rRNA contamination. ZymoResearch was kind enough…
Trimming - Additional 20bp from C.virginica Gonad RNAseq with fastp on Mox
2022
Miscellaneous
When I previously aligned trimmed RNAseq reads to the NCBI C.virginica genome (GCF_002022765.2) on 20210726, I specifically noted that alignment rates were consistently lower for males than females. However, I let that discrepancy distract me from a the larger issue: low…
Transcriptome Assembly - Genome-guided C.virginica Adult Gonad OA RNAseq Using Trinity on Mox
2022
Miscellaneous
As part of this project, Steven’s asked that I identify long, non-coding RNAs (lncRNAs) (GitHub Issue) in the Crassostrea virginica (Eastern oyster) adult OA gonad RNAseq data we have. The initial step for this is to assemble transcriptome. I generated…
RNAseq Alignment - C.virginica Adult OA Gonad Data to GCF_002022765.2 Genome Using HISAT2 on Mox
2022
Miscellaneous
As part of this project, Steven’s asked that I identify long, non-coding RNAs (lncRNAs) (GitHub Issue) in the Crassostrea virginica (Eastern oyster) adult OA gonad RNAseq data we have. The initial step for this is to assemble…
Data Wrangling - C.virginica Gonad RNAseq Transcript Counts Per Gene Per Sample Using Ballgown
2022
Miscellaneous
As we continue to work on the analysis of impacts of OA on Crassostrea virginica (Eastern oyster) gonads via DNA methylation and RNAseq (GitHub repo), we decided to compare the number of transcripts expressed per…
RNA Isolation - O.nerka Berdahl Tissues
2021
Miscellaneous
Finally got around to tackling this GitHub issue regarding isolating RNA from some Oncorhynchus nerka (sockeye salmon) tissues we have from Andrew Berdahl’s lab (a UW SAFS professor) to use for RNAseq and/or qPCR. We have blood, brain, gonad, and liver samples from individual salmon from two different groups: territorial and social…
SNP Characterization - C.bairdi v3.1 Transcriptome Assembly and Day2 DEG Pooled Samples RNAseq Data
2021
Tanner Crab RNAseq
I previously identified variants across the four Day 2 DEG pooled RNAseq samples (380822, 380823, 380824, 380825) on 20210909 within the
cbai_transcriptome_v3.1 transcriptome assembly. Now, I needed to actually do some analysis on the SNPs for the…
RNAseq Alignments - C.bairdi Day 2 Infected-Uninfected Temperature Increase-Decrease RNAseq to cbai_transcriptome_v3.1.fasta with Hisat2 on Mox
2021
Tanner Crab RNAseq
Ealier today, I created the necessary Hisat2 index files for
cbai_transcriptome_v3.1. Next, I needed to actually get the alignments run. The alignments were performed using HIS…
Assembly Indexing - C.bairdi Transcriptome cbai_transcriptome_v3.1.fasta with Hisat2 on Mox
2021
Tanner Crab RNAseq
…
Transcript Identification and Quantification - C.virginia RNAseq With NCBI Genome GCF_002022765.2 Using StringTie on Mox
2021
Miscellaneous
After having run
HISAT2 to index and identify exons and splice sites in the NCBI Crassostrea virginica (Eastern oyster) genome (GCF_002022765.2) on 20210720, the next step was to identify and quantify transcripts from…Genome Assembly - Olurida_v090 with BGI Illumina and PacBio Hybrid Using Wengan on Mox
2021
Olympia Oyster Genome Assembly
I was recently tasked with adding annotations for our Ostrea lurida genome assembly to NCBI. As it turns out, adding just annotation files can’t be done since the genome was initially…
Transcriptome Assessment - BUSCO Metazoa on C.bairdi Transcriptome v4.0 on Mox
2021
Tanner Crab RNAseq
I previously created a C.bairdi de novo transcriptome assembly v4.0 with Trinity from all our C.bairdi RNAseq reads which had BLASTx matches to the C.opilio genome and decided to assess its “completeness” using…
DIAMOND BLASTx - C.bairdi RNAseq vs C.opilio Genome Proteins on Mox
2021
Tanner Crab RNAseq
We want to generate an additional Tanner crab (Chionoecetes bairdi) transcriptome, per this GitHub issue, to generate an additional C.bairdi transcriptome. This has come about due to the release of the genome of a very closely…
Data Wrangling - Gene ID Extraction from P.generosa Genome GFF Using Methylation Machinery Gene IDs
2021
Miscellaneous
Per this GitHub issue, Steven provided a list of methylation-related gene names and wanted to extract the corresponding Panopea generosa ([Pacific geoduck (Panopea generosa)](http://en.wikipedia.org/wiki/Geodu…
Data Wrangling - Gene ID Extraction from P.generosa Genome GFF Using Methylation Machinery List
2021
Miscellaneous
Per this GitHub Issue Steven asked that I take a list of gene names associated with DNA methylation and see if I could extract a list of Panopea generosa (Panopea generosa) gene IDs and corresponding BLAST e-values for each from…
Data Received - Anthopleura elegantissima - aggregating anenome - NanoPore Genome Sequence from Jay Dimond
2021
Miscellaneous
Jay asked me to help get his A.elegantissima (aggregating anenome) NanoPore gDNA sequencing data submitted to NCBI Sequencing Read Archive (SRA). He sent a hard drive (HDD) with all the NanoPore sequencing Fast5 files. The HDD was received on 2/2/2021. Here’re are details provided in the…
Alignments - C.bairdi RNAseq Transcriptome Alignments Using Bowtie2 on Mox
2020
Tanner Crab RNAseq
I had previously attempted to compare all of our C.bairdi transcriptome assemblies using DETONATE on 20200601, but, due to hitting time limits on Mox, failed to successfully get the analysis to complete. I realized that the limiting factor was performing FastQ alignments, so I…
Trimming - Haws Lab C.gigas Ploidy pH WGBS 10bp 5 and 3 Prime Ends Using fastp and MultiQC on Mox
2020
Miscellaneous
Making the assumption that the 24 C.gigas ploidy pH WGBS data we receved 20201205 will be analyzed using
Bismark, I decided to go ahead and trim the files according to Bismark guidelines…
Data Received - C.gigas Diploid-Triploid pH Treatments Ctenidia WGBS from ZymoResearch
2020
Data Received
Today we received the whole genome bisulfite sequencing (WGBS) from the 24 C.gigas diploid-triploid samples subjected to different pH that were submitted 20200824. The lengthy turnaround time was due to a bad lot of reagents, which forced them Zymo to find a different manufacturer in order to…
Sample Submission - M.magister MBD BSseq Libraries for MiSeq at NOAA
2020
Samples Submitted
Earlier today I quantified the libraries with the Qubit in preparation for sample pooling and sequencing. Before performing a full sequencing run, Mac wanted to select a subset of the libraries based on the experimental…
MBD BSseq Library Prep - M.magister MBD-selected DNA Using Pico Methyl-Seq Kit
2020
Miscellaneous
After finishing the final set of eight MBD selections on 20201103, I’m finally ready to make the BSseq libraries using the Pico Methyl-Seq Library Prep Kit (ZymoResearch) (PDF). I followed the manufacturer’s protocols with the following notes/changes (organized by each section in the…
Transcriptome Assessment - Crustacean Transcripome Completeness Evaluation Using BUSCO on Mox
2020
Miscellaneous
Grace was recently working on writing up a manuscript which did a basic comparison of our C.bairdi transcriptome (
cbai_transcriptome_v3.1) (see the Genomic Resources wiki for more deets) to two other species’ transcriptome assemblies. We wanted BUSCO evaluations as part of this comparison, but the two other species did not have BUSCO scores…
RNAseq Alignments - S.salar HISAT2 BAMs to GCF_000233375.1_ICSASG_v2_genomic.gtf Transcriptome Using StringTie on Mox
2020
Miscellaneous
This is a continuation of addressing Shelly Trigg’s (regarding some Salmo salar RNAseq data) request (GitHub Issue) to trim (completed 20201029), perform genome alignment (completed on…
MBD Selection - M.magister Sheared Gill gDNA 8 of 24 Samples Set 2 of 3
2020
Miscellaneous
Click here for notebook on the first eight samples processed. M.magister (Dungeness crab) gill gDNA provided by Mackenzie Gavery was previously sheared on 20201026 and three samples were subjected to additional rounds…
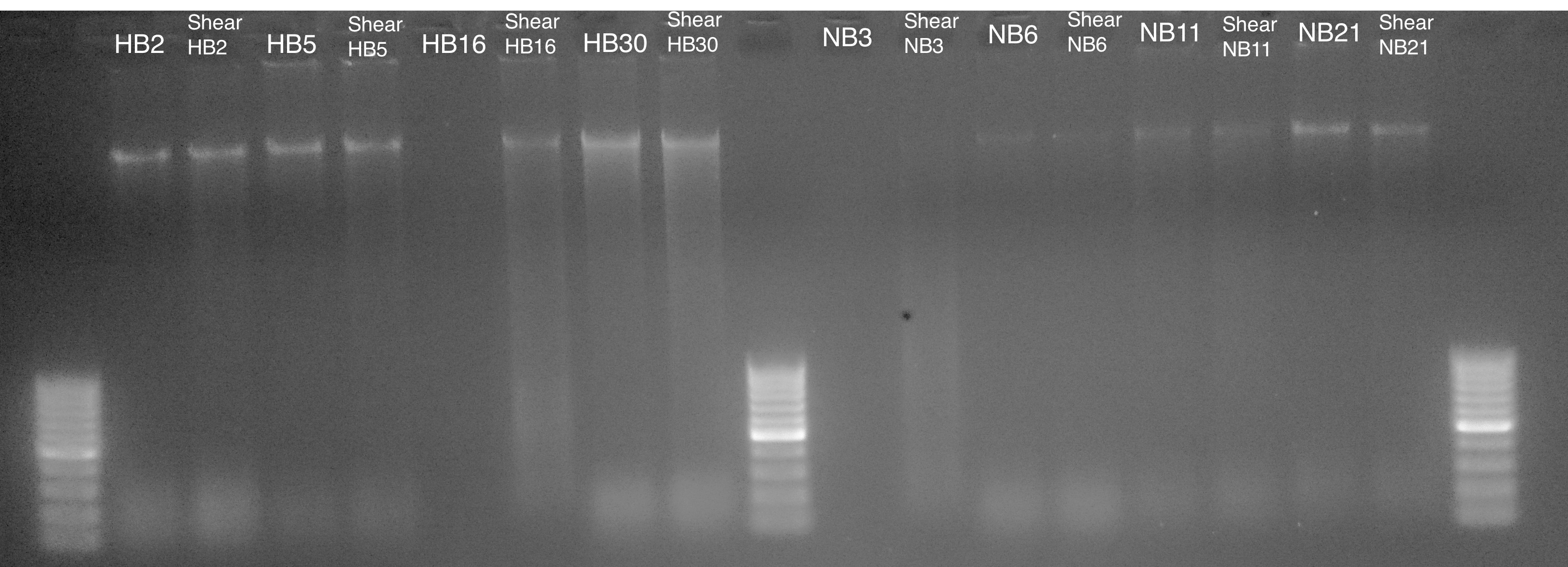
DNA Shearing - M.magister gDNA Shearing All Samples and Bioanalyzer
2020
Miscellaneous
I previously ran some shearing tests on 20201022 to determine how many cycles to run on the sonicator (Bioruptor 300; Diagenode) to achieve an average fragment length of ~350 - 500bp in preparation for MBD-BSseq. The…
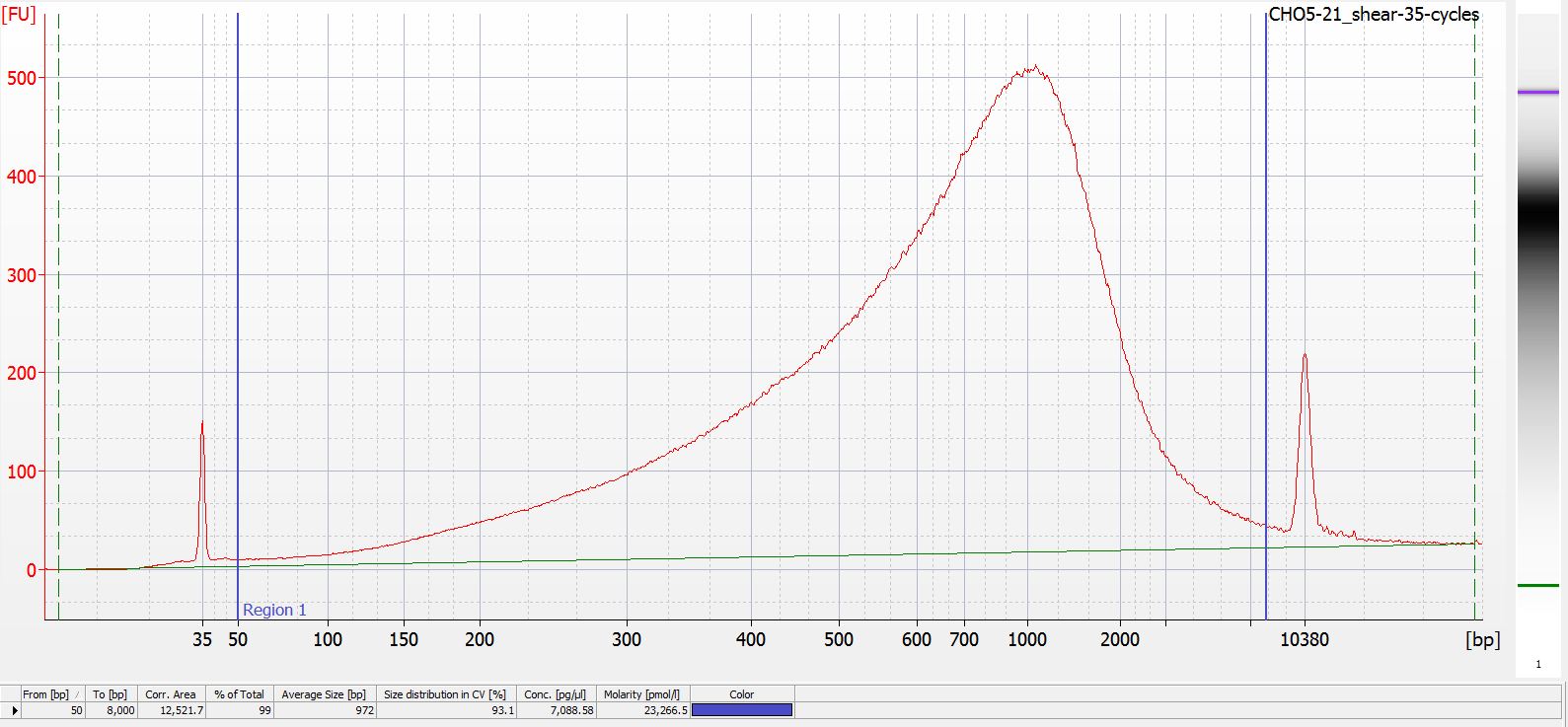
DNA Shearing - M.magister CH05-21 gDNA Full Shearing Test and Bioanalyzer
2020
Miscellaneous
Yesterday, I did some shearing of Metacarcinus magister gill gDNA on a test sample (CH05-21) to determine how many cycles to run on the sonicator (Bioruptor 300; Diagenode) to achieve an average fragment length of ~350 - 500bp in…
DNA Shearing - M.magister gDNA Shear Testing and Bioanalyzer
2020
Miscellaneous
Steven assigned me to do some MBD-BSseq library prep (GitHub Issue) for some Dungeness crab (Metacarcinus magister) DNA samples provided by Mackenzie Gavery. The DNA was isolated from juvenile (J6/J7 developmental stages) gill tissue. One of the first steps in MBD-BSseq is to fragment…
Read Mapping - C.bairdi 201002558-2729-Q7 and 6129-403-26-Q7 Taxa-Specific NanoPore Reads to cbai_genome_v1.01.fasta Using Minimap2 on Mox
2020
Miscellaneous
After extracting FastQ reads using
seqtk on 20201013 from the various taxa I had been interested in, the next thing needed doing was mapping reads to the cbai_genome_v1.01 “genome” assembly from…
Taxonomic Assignments - C.bairdi 6129-403-26-Q7 NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on emu
2020
Miscellaneous
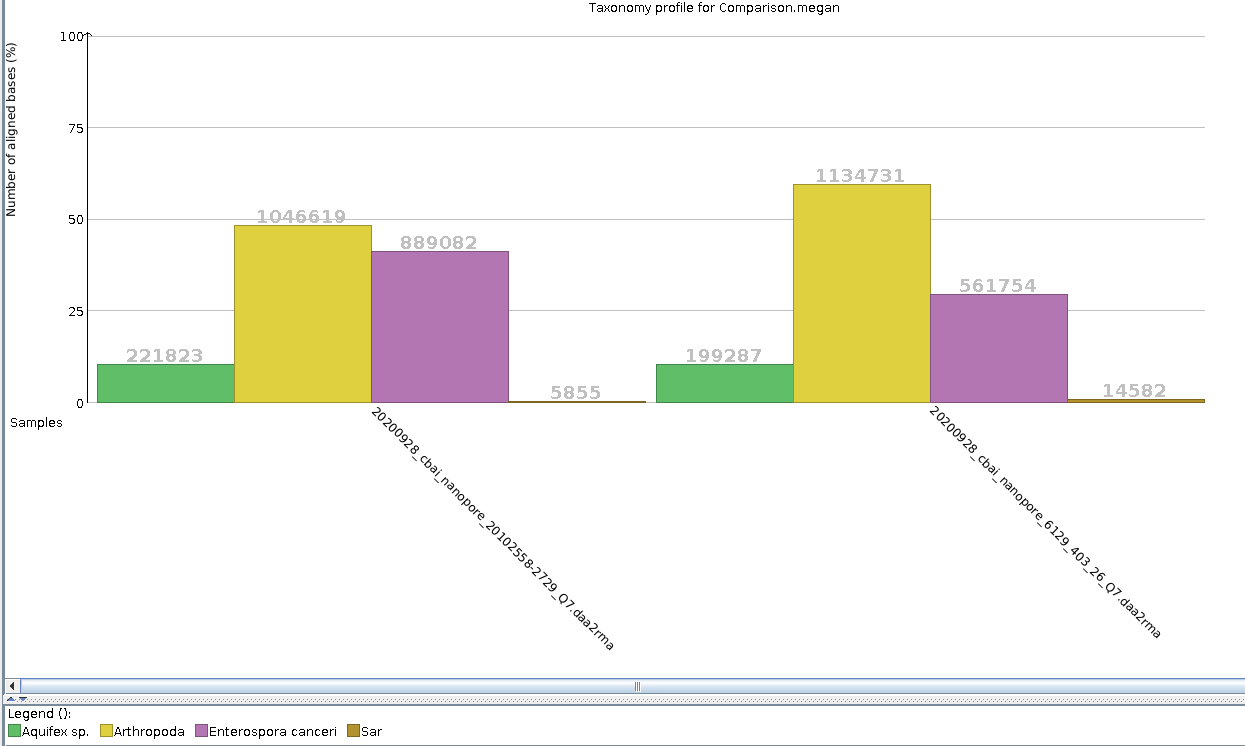
After noticing that the initial MEGAN6 taxonomic assignments for our combined C.bairdi NanoPore data from 20200917 revealed a high number of bases assigned to E.canceri and Aquifex…
Taxonomic Assignments - C.bairdi 20102558-2729-Q7 NanoPore Reads Using DIAMOND BLASTx on Mox and MEGAN6 daa2rma on emu
2020
Miscellaneous
After noticing that the initial MEGAN6 taxonomic assignments for our combined C.bairdi NanoPore data from 20200917 revealed a high number of bases assigned to E.canceri and Aquifex…
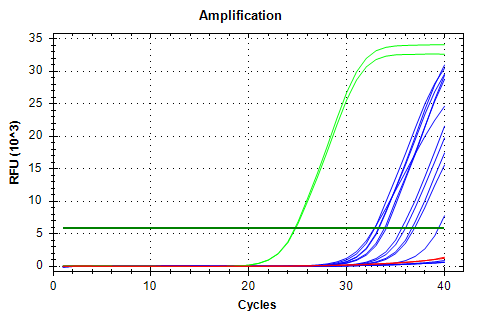
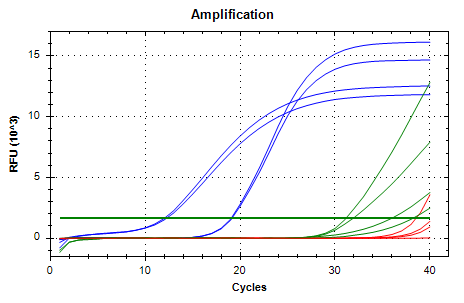
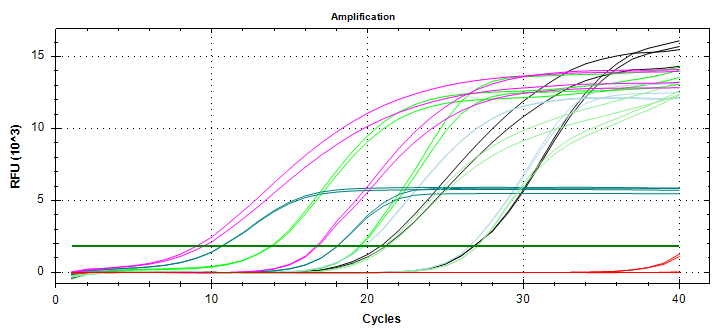
qPCR - Geoduck Normalizing Gene Primers 28s-v4 and EF1a-v1 Tests
2020
Miscellaneous
On Monday (20200914), I checked a set of 28s and EF1a primer sets and determined that 28s-v4 and EF1a-v1 were probably the best of the bunch, although they all looked great. So, I needed to test these out on some individual cDNA samples…
Samples Submission - Supplemental Ronits C.gigas Diploid-Triploid Ctendidia gDNA for WGBS by ZymoResearch
2020
Samples Submitted
I re-quantified samples for Zymo whole genome bisulfite sequencing (WGBS) project 3534 earlier today due to ZymoResearch indicating there was insufficient DNA for most of the samples and submitted an additional 1ug of each…
DNA Quantification - Re-quant Ronits C.gigas Diploid-Triploid Ctenidia gDNA Submitted to ZymoResearch
2020
Miscellaneous
I received notice from ZymoResearch yesterday afternoon that the DNA we sent on 20200820 for this project (Quote 3534) had insufficient DNA for sequencing for most of the samples. This was, honestly, shocking. I had even submitted well over the minimum amount of DNA required (submitted…
Primer Design and In-Silico Testing - Geoduck Reproduction Primers
2020
Miscellaneous
Shelly asked that I re-run the primer design pipeline that Kaitlyn had previously run to design a set of reproduction-related qPCR primers. Unfortunately, Kaitlyn’s Jupyter Notebook wasn’t backed up and she accidentally…
SRA Submission - Geoduck Epigenetic Ocean Acidification RNAseq
2020
SRA Submission
Can’t remember where it was discussed (probably lab meeting), but I created a GitHub Issue to add all of geoduck RNAseq data to NCBI Short Read Archive (SRA). Anyway, got all the remaining RNAseq data uploaded to the NCBI SRA and organized into the correct…
ENA Submission - Ostrea lurida draft genome Olurida_v081.fa
2020
Olympia Oyster Genome Sequencing
Submitted our Ostrea lurida v081 genome assembly FastA to the European Nucloetide Archive.
Transcriptome Annotation - Trinotate C.bairdi Transcriptome-v1.7 on Mox
2020
Miscellaneous
After creating a de novo assembly of C.bairdi transcriptome v1.7 on 20200527, performing BLASTx annotation on 202000527, and TransDecoder for ORF identification on 20200527, I continued the annotation process by…
Transcriptome Comparisons - C.bairdi BUSCO Scores
2020
Miscellaneous
Since we’ve generated a number of versions of the C.bairdi transcriptome, we’ve decided to compare them using various metrics. Here, I’ve compared the BUSCO scores generated for each transcriptome using BUSCO’s built-in plotting script. The…
Transcriptome Assembly - C.bairdi All Pooled Arthropoda-only RNAseq Data with Trinity on Mox
2020
Miscellaneous
For completeness sake, I wanted to create an additional C.bairdi transcriptome assembly that consisted of Arthropoda only sequences from just pooled RNAseq data (since I recently generated a similar…
SRA Library Assessment - Determine RNAseq Library Strandedness from P.trituberculatus SRA BioProject PRJNA597187
2020
Miscellaneous
We’ve produced a number of C.bairid transcriptomes utilizing different assembly approaches (e.g. Arthropoda reads only, stranded libraries only, mixed strandedness libraries…
Transcriptome Annotation - Trinotate C.bairdi Transcriptome-v1.6 on Mox
2020
Miscellaneous
After creating a de novo assembly of C.bairdi transcriptome v1.6 on 20200518, performing BLASTx annotation on 202000519, and TransDecoder for ORF identification on 20200519, I continued the annotation process by…
Transcriptome Annotation - Trinotate C.bairdi Transcriptome v2.0 from 20200502 on Mox
2020
Miscellaneous
After performing de novo assembly on all of our Tanner crab RNAseq data (no taxonomic filter applied, either) on 20200502 and performing BLASTx annotation on 20200508, I continued the annotation process…
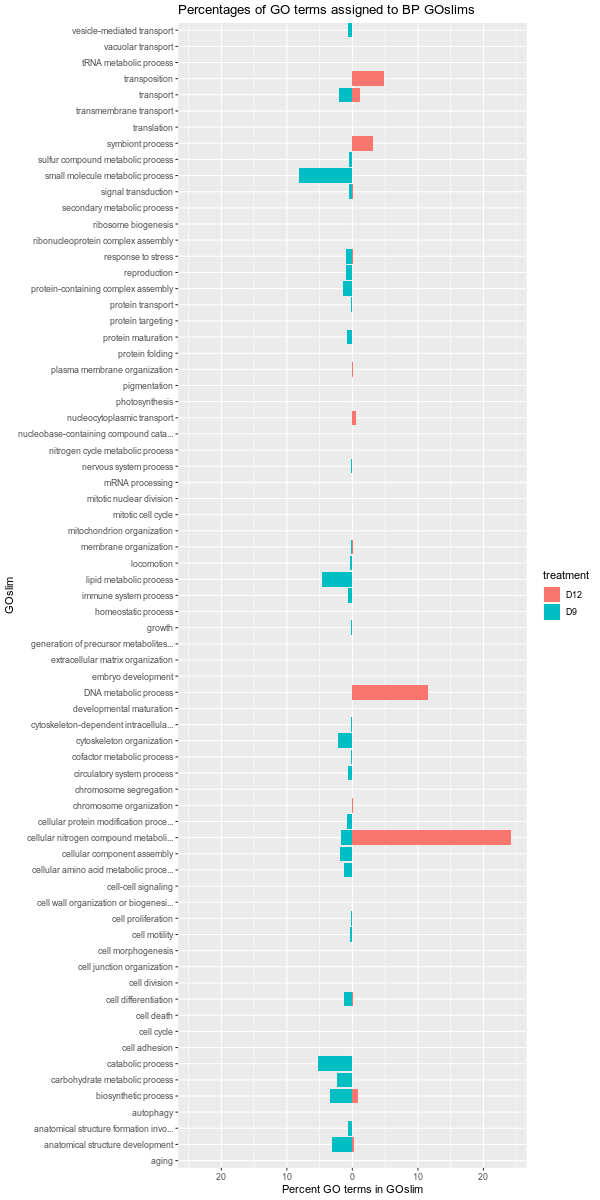
GO to GOslim - C.bairdi Enriched GO Terms from 20200422 DEGs
2020
Miscellaneous
After running pairwise comparisons and identify differentially expressed genes (DEGs) on 20200422 and finding enriched gene ontology terms, I decided to map the GO terms to Biological Process GOslims. Additionally, I decided to try another level of comparison (I’m not…
Transcript Abundance - C.bairdi Alignment-free with Salmon on Mox for Grace
2020
Miscellaneous
Per this GitHub Issue, Grace and Steven asked if I could help by generating a transcript abundance file for Grace to use with EdgeR. To do so, I used Salmon for alignment-free transcript abundance estimates due to its speed and…
Data Received - C.bairdi RNAseq Data from Genewiz
2020
Data Received
Tanner Crab RNAseq
We received the RNAseq data from the RNA that was sent out by Grace on 20200212.
NanoPore Sequencing - C.bairdi gDNA 6129_403_26
2020
Miscellaneous
After getting high quality gDNA from Hematodinium-infected C.bairdi hemolymph on 2020210 we decided to run some of the sample on the NanoPore MinION, since the flowcells have a very short shelf life. Additionally, the results from this will also help inform us…
qPCR - C.bairdi RNA Check for Residual gDNA
2020
Tanner Crab RNAseq
Previuosly checked existing crab RNA for residual gDNA on 20200226 and identified samples with yields that were likely too low, as well as samples with residual gDNA. For those samples, was faster/easier to just isolate more RNA and…
Trimming/MultiQC - Methcompare Bisulfite FastQs with fastp on Mox
2020
Miscellaneous
Steven asked me to trim a set of FastQ files, provided by Hollie Putnam, in preparation for methylation analysis using Bismark. The analysis is part of a coral project comparing DNA methylation profiles of different species, as well as comparing different sample…
RNA Isolation and Quantification - C.bairdi RNA from Hemolymph Pellets in RNAlater
2020
Tanner Crab RNAseq
Based on qPCR results testing for residual gDNA from 20200225, a set of 24 samples were identified that required DNase treatment and/or additional RNA. I opted to just isolate more RNA from all samples, since the kit includes a…
DNA Isolation & Quantification - Additional C.bairdi gDNA from Sample 6129_403_26
2020
Miscellaneous
Earlier today I isolated gDNA from C.bairi 6129_403_26 hemolymph pellets and recovered decently intact gDNA that could be used for sequencing. However, I still need more gDNA, so will isolate that (and co-isolate RNA, since I’m going through the…
Gene Expression - Hematodinium MEGAN6 with Trinity and EdgeR
2020
Miscellaneous
After completing annotation of the Hematodinium MEGAN6 taxonomic-specific Trinity assembly using Trinotate on 20200126, I performed differential gene expression analysis and gene ontology (GO) term enrichment analysis using Trinity’s scripts to run EdgeR and GOseq, respectively. The…
Gene Expression - C.bairdi MEGAN6 with Trinity and EdgeR
2020
Tanner Crab RNAseq
After completing annotation of the C.bairdi MEGAN6 taxonomic-specific Trinity assembly using Trinotate on 20200126, I performed differential gene expression analysis and gene ontology (GO) term enrichment analysis using Trinity’s scripts to run EdgeR and GOseq, respectively, across all…
RNA Isolation and Quantification - C.bairdi Hemocyte Pellets in RNAlater Troubleshooting
2020
Tanner Crab RNAseq
After the failure to obtain RNA from any C.bairdi hemocytes pellets (out of 24 samples processed) on 20200117, I decided to isolate RNA from just a subset of that group to determine if I screwed something up last time or something. Also, I am testing two different preparations of…
Data Wrangling - Arthropoda and Alveolata Taxonomic RNAseq FastQ Extractions
2020
Tanner Crab RNAseq
After using MEGAN6 to extract Arthropoda and Alveolata reads from our RNAseq data on 20200114 (for reference, these include RNAseq data using a newly established “shorthand”: 2018, 2019), I realized that the FastA…
RNAseq Reads Extractions - C.bairdi Taxonomic Reads Extractions with MEGAN6 on swoose
2020
Tanner Crab RNAseq
I previously ran BLASTx and “meganized” the output DAA files on 20200103 (for reference, these include RNAseq data using a newly established “shorthand”: 2018, 2019) and now need to use MEGAN6 to bin the results into the proper…
NanoPore Sequencing - Initial NanoPore MinION Lambda Sequencing Test
2020
Miscellaneous
We recently acquired a NanoPore MinION sequencer, FLO-MIN106 flow cell and the Rapid Sequencing Kit (SQK-RAD004). The NanoPore website provides a pretty thorough an user-friendly walk-through of how to begin using the system for the first time. With that said, I believe the user needs…
Trimming/FastQC/MultiQC - C.bairdi RNAseq FastQ with fastp on Mox
2019
Tanner Crab RNAseq
Grace/Steven asked me to generate a de novo transcriptome assembly of our current C.bairdi RNAseq data in this GitHub issue. As part of that, I needed to quality trim the data first. Although I could automate this as part of the transcriptome assembly (Trinity has Trimmomatic built-in), I would…
Samples Received - C.bairdi Hemolymph and Tissue in Ethanol from Pam Jensen
2019
Samples Received
Pam…
Data Wrangling - Rename Pgenerosa_v074 Bismark Coverage Files Scaffold Names
2019
Geoduck Genome Sequencing
After a meeting today, Steven and Hollie realized that the Bismark coverage files were still using the old scaffold names, stemming from the Pgenerosa_v074 naming. I’d previously updated filenames and…
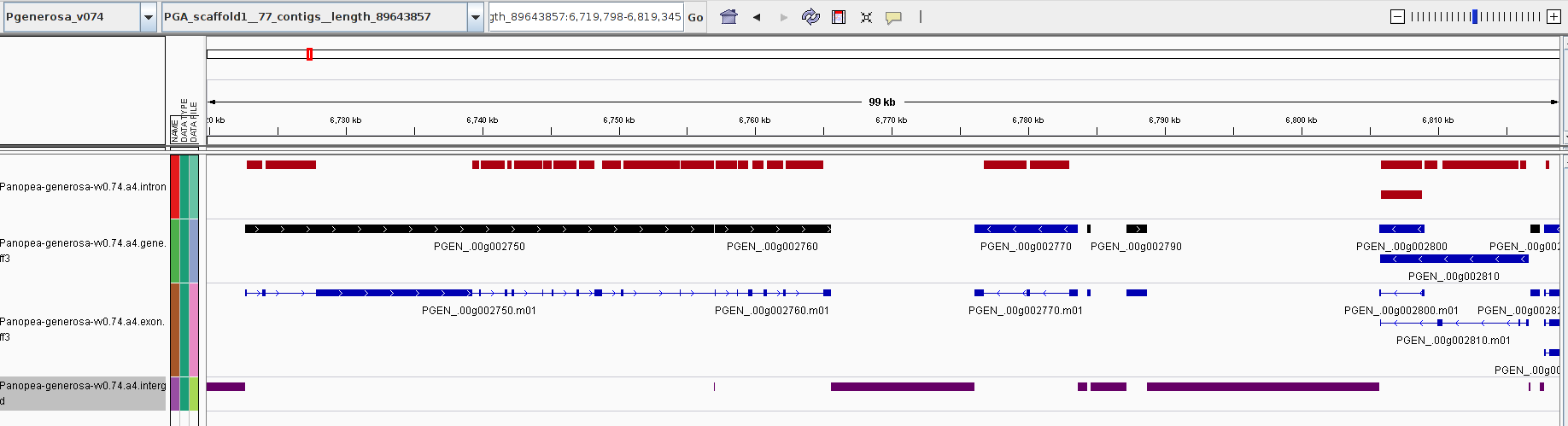
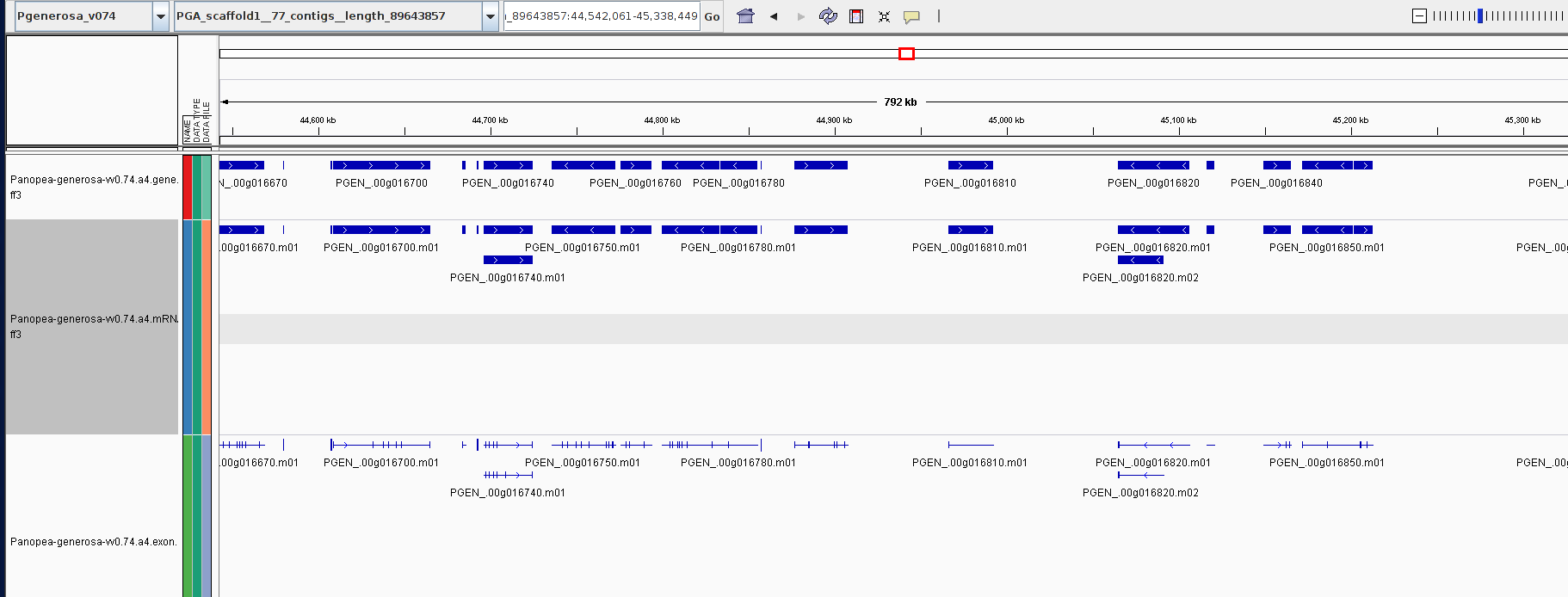
Genome Feature Counts - Panopea-generosa-vv0.74.a4
2019
Geoduck Genome Sequencing
Due to a bug with GenSAS, the final GFF generated by their system to did not properly merge all of the repeats data properly. The repeats data below should be ignored! It appears that the other features data (e.g. CDS, exons, etc) was unaffected…
Data Received - C.bairdi RNAseq Day9-12-26 Infected-Uninfected
2019
Tanner Crab RNAseq
Data Received
Previously, we “received” this data, but it turns out it was incomplete (see 20191003).
Genome Comparison - Pgenerosa_v074 vs Pgenerosa_v070 with MUMmer Promer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs S.glomerata NCBI with MUMmer Promer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs M.yessoensis NCBI with MUMmer Promer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs H.sapiens NCBI with MUMmer Promer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs C.virginica NCBI with MUMmer Promer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs C.gigas NCBI with MUMmer Promer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs Pgenerosa_v070 with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs Pgenerosa_v074 with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs S.glomerata NCBI with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs M.yessoensis NCBI with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs C.gigas NCBI with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs H.sapiens NCBI with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
Genome Comparison - Pgenerosa_v074 vs C.virginica NCBI with MUMmer on Mox
2019
Miscellaneous
In continuing to further improve our geoduck genome annotation, I’m attempting to figure out why Scaffold 1 of our assembly doesn’t have any annotations. As part of that I’ve decided to perform a series of genome comparisons and see how they match up, with an…
RepeatMasker - Pgenerosa_v070 for Transposable Element ID on Roadrunner
2019
Geoduck Genome Sequencing
Continuing our various attempts at annotating our geoduck genome assemblies, I will be re-annotating our Pgenerosa_v070 (see Genome Resources GitHub wiki for deets) and realized I hadn’t run RepeatMasker on this assembly previously. Running RepeatMasker will generate a GFF that I can supply to MAKER to aid in repeats…
Genome Annotation - Pgenerosa_v074 Transcript Isoform ID with Stringtie on Mox
2019
Geoduck Genome Sequencing
After annotating Pgenerosa_v070 and comparing feature counts, there was a drastic difference between the two genome versions. Additionally, both of those genomes ended up with no CDS/exon/gene/mRNA features identified in the largest…
Genome Annotation - Pgenerosa_v070 Transcript Isoform ID with Stringtie on Mox
2019
Geoduck Genome Sequencing
After annotating Pgenerosa_v074 and comparing feature counts, there was a drastic difference between the two genome versions. Additionally, both of those genomes ended up with no CDS/exon/gene/mRNA features identified in the largest…
Genome Annotation - Pgenerosa_v070 Hisat2 Transcript Isoform Index
2019
Geoduck Genome Sequencing
This is the first step in getting transcript isoform…
Genome Annotation - Pgenerosa_v070 and v074 Top 18 Scaffolds Feature Count Comparisons
2019
Geoduck Genome Sequencing
After annotating Pgenerosa_v074 on 20190701, we noticed a large discrepancy in the number of transcripts that MAKER identified, compared to Pgenerosa_v070. As a reminder, the Pgenerosa_v074 is a subset of…
Genome Annotation - O.lurida 20190709-v081 Transcript Isoform ID with Stringtie on Mox
2019
Olympia Oyster Genome Sequencing
Earlier today, I generated the necessary Hista2 index, which incorporated splice sites and exons, for use with Stringtie in order to identify transcript isoforms in our 20190709-Olurida_v081…
Genome Assessment - BUSCO Metazoa on Pgenerosa_v074 on Mox
2019
Geoduck Genome Sequencing
Ran BUSCO on Mox for our Pgenerosa_v74 genome assembly to assess…
Genome Annotation - Olurida_v081 with MAKER and Tissue-specific Transcriptomes on Mox
2019
Olympia Oyster Genome Sequencing
I previously annotated our Olurida_v081 genome with MAKER using our “canonical” transcriptome, Olurida_transcriptome_v3.fasta as the EST evidence utilized by MAKER. A discussion on one of our Slack channels related to the lack of isoform…
Genome Annotation - O.lurida (v081) Transcript Isoform ID with Stringtie on Mox
2019
Olympia Oyster Genome Sequencing
Earlier today, I generated the necessary Hista2 index, which incorporated splice sites and exons, for use with Stringtie in order to identify transcript isoforms in our Olurida_v081…
Sample Submission - Tanner Crab Infected vs Uninfected RNAseq
2019
Tanner Crab RNAseq
Samples Submitted
After reviewing our options for sample pooling, we decided to do a comparison of Infected vs. Uninfected crabs.
RNA Isolation and Quantification - C.bairdi Hemolymph Pellet in RNAlater
2019
Miscellaneous
After the success we had isolating RNA using the Quick-DNA/RNA Microprep Plus Kit (ZymoResearch), Steven had me isolate RNA from a list of ~117 samples. Of that list, I was able to find 66 crab hemolymph pelleted RNAlater samples. The “missing” samples were most likely…
Data Analysis - C.virginica MBD Sequencing Coverage
2019
Miscellaneous
A while ago, Steven tasked me with assessing some questions related to the sequencing coverage we get doing MBD-BSseq in this GitHub issue. At the heart of it all was really to try to get an idea of how much usable data we actually get when we do an MBD-BSseq project. Yaamini happened to have done an MBD-BSseq…
Metagenome Assemblies - P.generosa Water Samples Trimmed HiSeqX Data Using Megahit on Mox to Compare pH Treatments
2019
Miscellaneous
A report involving our work on the geoduck water metagenomics is due later this week and our in-depth analysis for this project using Anvi’o is likely to require at least another…
RNA Isolation and Quantification - Crab Hemolypmh Using Quick-DNA-RNA Microprep Plus Kit
2019
Tanner Crab RNAseq
In the continuing struggle to isolate RNA from the Chionoecetes bairdi hemolymph preserved in RNAlater, Pam managed to find the Quick-DNA-RNA Microprep Plus Kit (ZymoResearch) as a potential option. We received a free sample of the kit and so…
Transcriptome Assembly - Geoduck Tissue-specific Assembly Larvae Day5 EPI99 with HiSeq and NovaSeq Data on Mox
2019
Miscellaneous
I previously assembled and annotated P.generosa larval Day 5 transcriptome (20190318 - mislabeled as Juvenile Day 5 in my previous notebook entries) using just our HiSeq data from our Illumina collaboration. This was a an…
Transcriptome Assembly - Geoduck Tissue-specific Assembly Juvenile Ambient OA EPI123 with HiSeq Data on Mox
2019
Miscellaneous
Ran a de novo assembly on our HiSeq data from Hollie’s juvenile EPI 123 sample “ambient OA”. This was done for Christian to use in some long, non-coding RNA (lncRNA) analysis.
Sample Submission - Lotterhos C.virginica Mantle MBD DNA to ZymoResearch for BSseq
2019
Miscellaneous
Crassostrea virginica samples that were subjected to MBD enrichment (completed 20190319) were shipped, on dry ice, to ZymoResearch today for BSseq. They will perform bisulfite conversion, library prep, and…
Data Management - SRA Submission Panopea generosa Day 5 Larvae RNAseq
2019
Miscellaneous
In preparation for some manuscripts, Steven asked that I get some geoduck RNAseq data upload to the NCBI sequencing read archive (SRA) in this GitHub Issue. Specifically, he needed the data corresponding to day 5 (pos-fertilization) larve RNAseq data that was prepped/run by Illumina on the NovaSeq. Original sample…
MBD Enrichment - C.virginica Sheared Mantle DNA from 20190306
2019
Miscellaneous
Last week I performed MBD enrichment on 12 of the 24 Lotterhos C.viriginica mantle DNA on samples I had sheared on 20190306. I proceeded with MBD enrichment using the MethylMiner Kit (Invitrogen) on the 12 remaining samples of the 24. I followed the manufacturer’s protocol for…
DNA Shearing & Bioanalyzer - Lotterhos C.virginica Mantle gDNA from 2018114
2019
Miscellaneous
Crassostrea virginica mantle gDNA that had previously been isolated on 20181114 was evaluated for integrity via agaroase gel yesterday (20190305). Everything looked pretty good, so proceeded with shearing in…
Genome Assessment - BUSCO Metazoa on P.generosa v071 on Mox
2019
Miscellaneous
Ran BUSCO on our completed annotation of the P.generosa v071 genome (GFF) (subset of sequences >10kbp). See this notebook entry for genome annotation info. This provides a nice metric on how “complete” a genome assembly (or transcriptome) is. Additionally…
Genome Annotation - Pgenerosa_v070 MAKER on Mox
2019
Geoduck Genome Sequencing
Here it goes, a massive undertaking of attempting to annotate the Pgenerosa_v070 genome (FastA; 2.1GB) using MAKER on Mox! I previously started this on 20190115, but killed it in order to fix a number of different issues with the script that were causing problems. I decided the changes were substantial enough, that I’d just make a…
DNA Isolation and Quantification - Ronit’s C.gigas Ploidy Ctenidia
2019
Miscellaneous
Last week, I isolated DNA from all of Ronti’s ctenidia samples, however one sample (D18-C) didn’t isolate properly. So, I performed another isolation procedure with another section of frozen tissue. Tissue was excised from frozen…
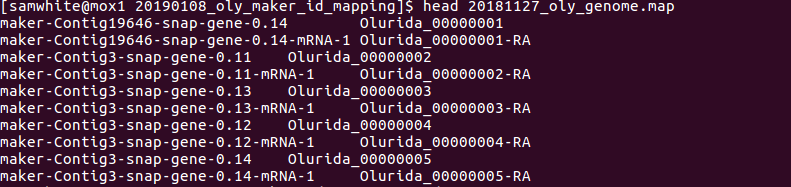
Annotation - Olurida_v081 MAKER ID Mapping
2019
Olympia Oyster Genome Sequencing
After getting through the initial MAKER annotation and SNAP gene predictions, I needed (wanted?) to simplify…
FastQC and Trimming - Metagenomics (Geoduck) HiSeqX Reads from 20180809
2018
Miscellaneous
Steven tasked me with assembling our geoduck metagenomics HiSeqX data. The first part of the process is examining the quality of the sequencing reads, performing quality trimming, and then checking the quality of the trimmed reads. It’s also…
Annotation - Olurida_v081 MAKER on Mox
2018
Olympia Oyster Genome Sequencing
Remarkably, I managed to burn through our Xsede computing resources and don’t have terribly much to show…
SRA Submission - Olymia oyster Whole Genome BS-seq Data
2018
Olympia oyster reciprocal transplant
Submitted our whole genome bisulfite sequencing data to NCBI Sequence Read Archive (SRA).
TrimGalore/FastQC/MultiQC - C.virginica Oil Spill MBDseq Concatenated Sequences
2018
LSU C.virginica Oil Spill MBD BS Sequencing
Previously concatenated and analyzed our Crassostrea virginica oil spill MBDseq data with FastQC.
Genome Annotation – Olympia oyster genome annotation results #02
2018
Olympia Oyster Genome Sequencing
Yesterday, I annotated our Olympia oyster genome using WQ-MAKER in just 7hrs!.
Genome Annotation - Olympia oyster genome annotation results #01
2018
Olympia Oyster Genome Sequencing
Yesterday, I annotated our Olympia oyster genome using WQ-MAKER in just 7hrs!.
Mox – Over quota: Olympia oyster genome annotation progress (using Maker 2.31.10)
2018
Olympia Oyster Genome Sequencing
Per this GitHub Issue, Steven has cleared out files.
Mox - Olympia oyster genome annotation progress (using Maker 2.31.10)
2018
Olympia Oyster Genome Sequencing
I decided to spend some time to figure…
Ubuntu - Fix “No Video Signal” Issue on Emu/Roadrunner
2018
Computer Servicing
Both Apple Xserves (Emu/Roadrunner)…
BS-seq Mapping - Olympia oyster bisulfite sequencing: TrimGalore > FastQC > Bismark
2018
BS-seq Libraries for Sequencing at Genewiz
MBD Enrichment for Sequencing at ZymoResearch
Steven asked me to evaluate our methylation sequencing data sets for Olympia oyster.
Assembly – SparseAssembler (k 111) on Geoduck Sequence Data
2018
Geoduck Genome Sequencing
Continuing to try to find the best kmer setting to work with SparseAssemlber after the last attempt failed due to a kmer size that was too large (k 131; which happens to be outside the max kmer size [127] for SparseAssembler), I re-ran SparseAssembler with an arbitrarily…
Data Management - Geoduck Phase Genomics Hi-C Data
2018
Geoduck Genome Sequencing
Samples Received
We received sequencing/assembly data from Phase Genomics.
Data Management - SRA Submission LSU C.virginica Oil Spill MBD BS-seq Data
2018
LSU C.virginica Oil Spill MBD BS Sequencing
Submitted the Crassostrea virginica (Eastern oyster) MBD BS-seq data we received on 20150413 to NCBI Sequence Read Archive.
TrimGalore/FastQC/MultiQC - Trim 10bp 5’/3’ ends C.virginica MBD BS-seq FASTQ data
2018
Miscellaneous
Steven found out that the Bismarck documentation (Bismarck is the bisulfite aligner we use in our BS-seq pipeline) suggests trimming 10bp from both the 5’ and 3’ ends. Since this is the next step in our pipeline, we figured we should probably just follow their…
TrimGalore/FastQC/MultiQC - 2bp 3’ end Read 1s Trim C.virginica MBD BS-seq FASTQ data
2018
Miscellaneous
Earlier today, I ran TrimGalore/FastQC/MultiQC on the Crassostrea virginica MBD BS-seq data from ZymoResearch and hard trimmed the first 14bp from each read. Things looked better at the 5’ end, but the 3’ end of each of…
Genome Assembly - SparseAssembler Geoduck Genomic Data, kmer=101
2018
Geoduck Genome Sequencing
Assembly complete. See end of post for data locations.
Samples Submitted - C. virginica gDNA, MBD, and MspI to Qiagen
2018
Samples Submitted
Sent Crassostrea virginica s…
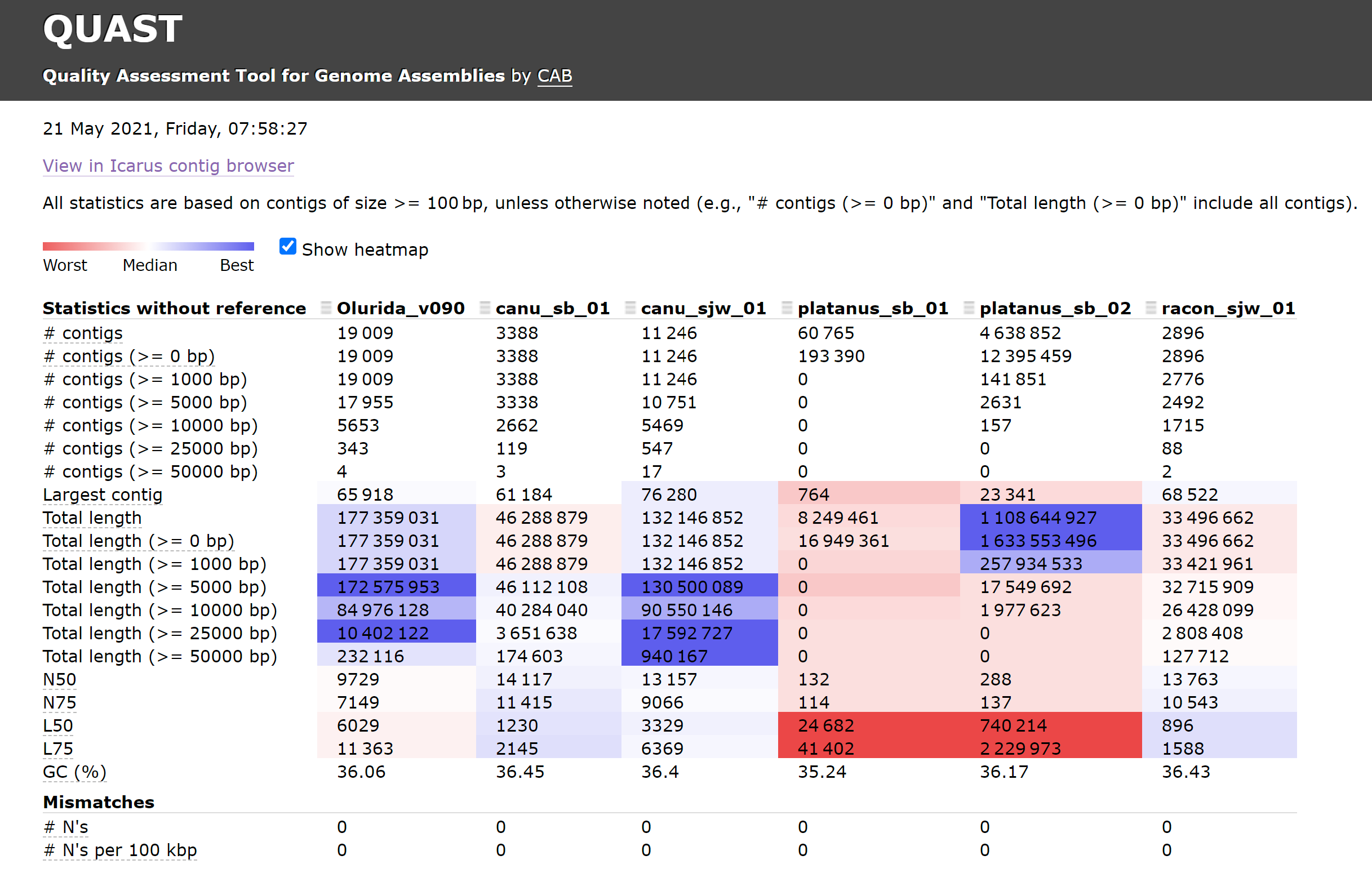
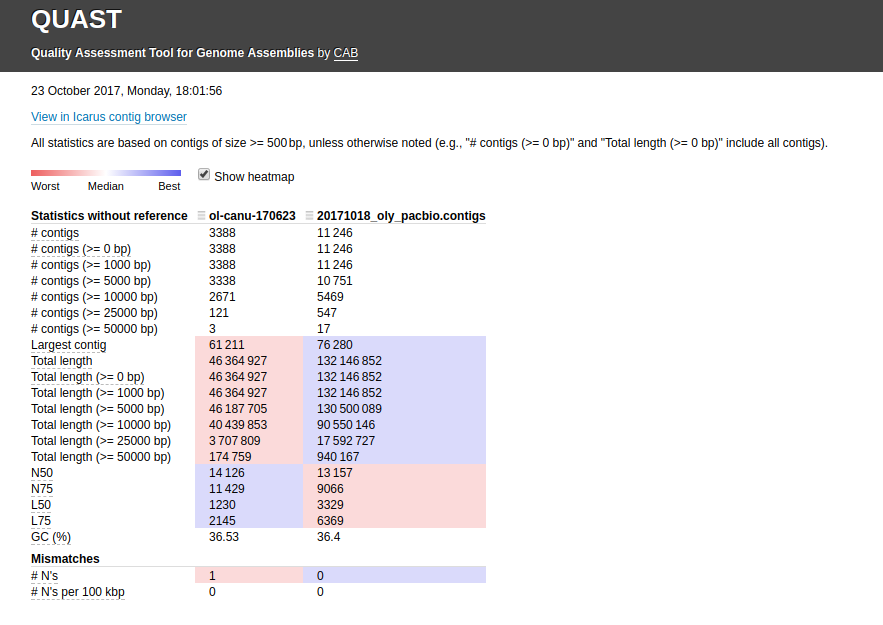
Assembly Comparisons – Oly Assemblies Using Quast
2018
Olympia Oyster Genome Sequencing
I ran Quast to compare all of our current Olympia oyster genome assemblies.
Samples Submitted - Pulverized Geoduck Tissues to Illumina for More 10x Genomics Sequencing
2017
Geoduck Genome Sequencing
Samples Submitted
Continuing Illumina’s generous efforts to use our geoduck samples to test out the robustness of their emerging sequencing technologies, they have requested we…
Genome Assembly – Olympia Oyster Illumina & PacBio Using PB Jelly w/BGI Scaffold Assembly
2017
Olympia Oyster Genome Sequencing
After another attempt to fix PB Jelly, I ran it again.
Assembly Comparison - Oly Assemblies Using Quast
2017
Olympia Oyster Genome Sequencing
I ran Quast to compare all of our current Olympia oyster genome assemblies.
Genome Assembly - Olympia oyster Illumina & PacBio Reads Using Redundans
2017
Olympia Oyster Genome Sequencing
Had problems with Docker and…
Genome Assembly - Olympia oyster Illumina & PacBio reads using MaSuRCA
2017
Olympia Oyster Genome Sequencing
Genome Assembly - Olympia Oyster Redundans with Illumina + PacBio
2017
Olympia Oyster Genome Sequencing
Redundans should assemble both Illumina and PacBio data, so let’s do that.
Genome Assembly - minimap/miniasm/racon Overview
2017
Olympia Oyster Genome Sequencing
Previously, I used the following three tools to do quick assembly of our Olympia oyster PacBio data:
Samples Submitted - Geoduck Ctenidia to Illumina for 10x Genomics Sequencing
2017
Geoduck Genome Sequencing
Samples Submitted
Continuing Illumina’s generous efforts to use our geoduck samples to test out the robustness of their emerging…
Project Progress - Olympia Oyster Genome Assemblies by Sean Bennett
2017
Olympia Oyster Genome Sequencing
Here’s a brief overview of what Sean has done with the Oly genome assembly front.
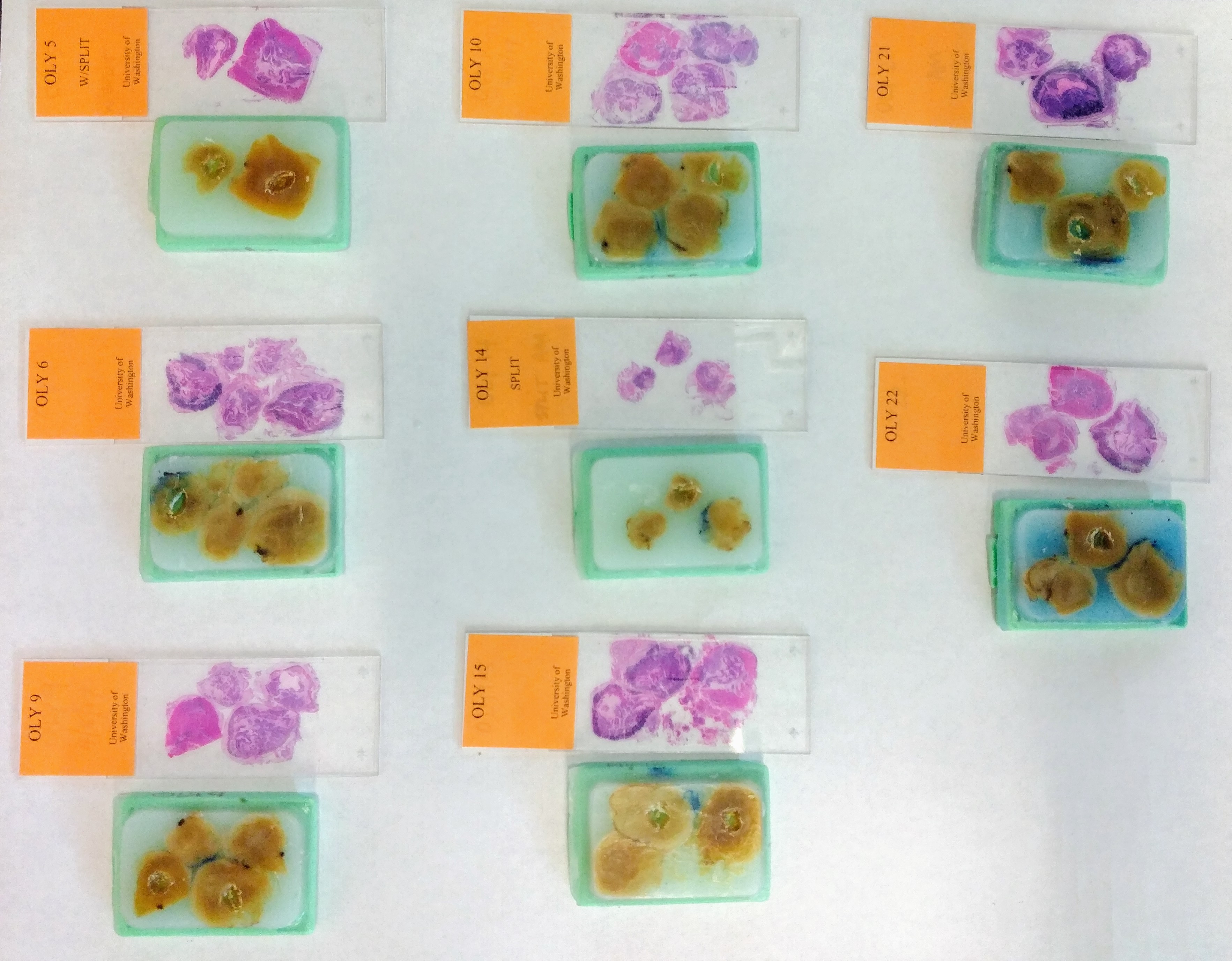
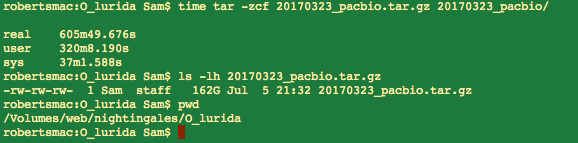
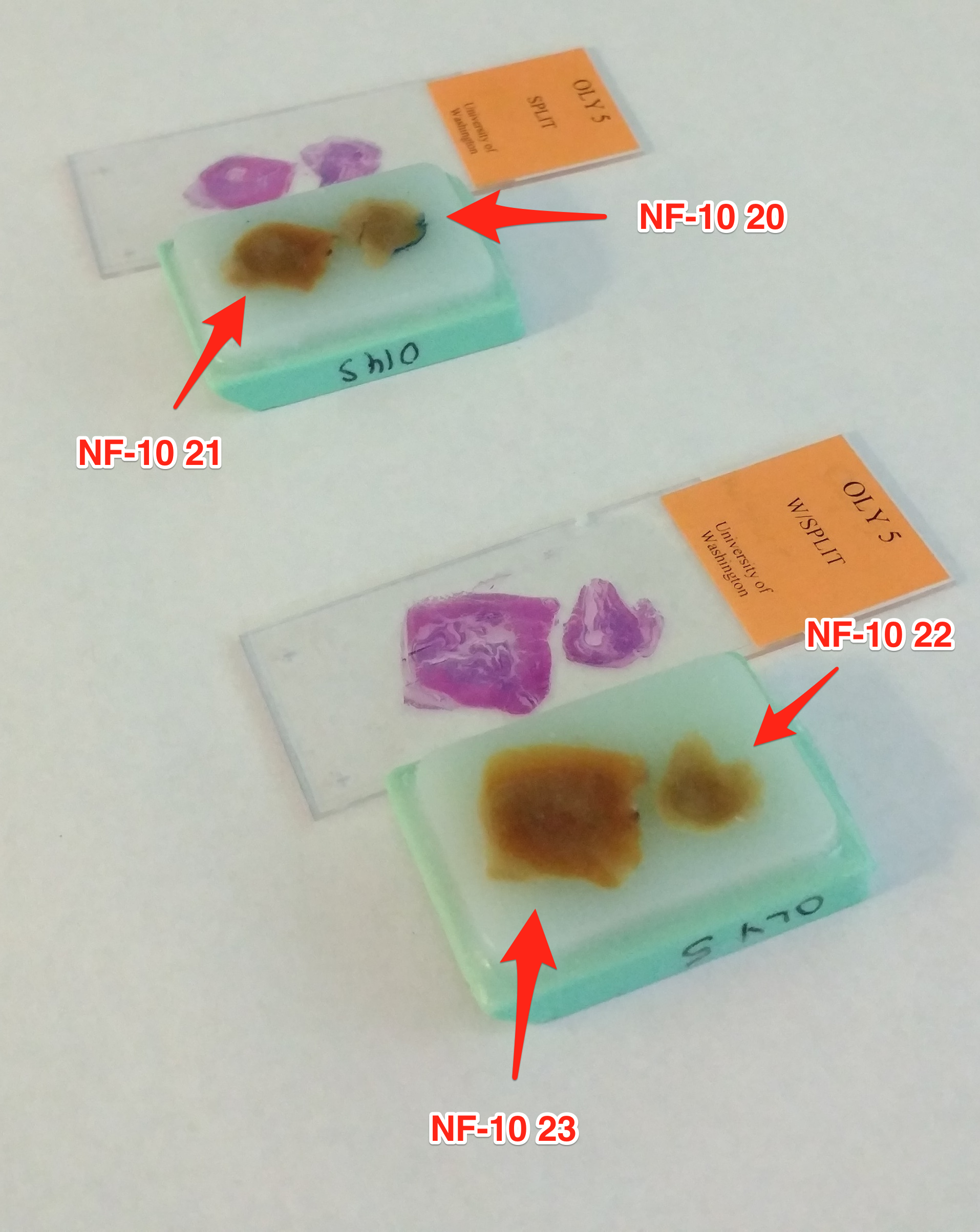
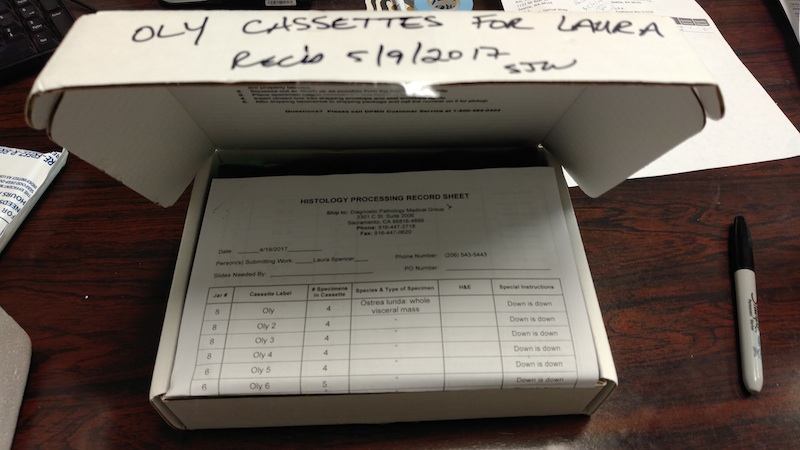
Data Management – Tarball of Olympia oyster UW PacBio Data from 20170323
2017
Olympia Oyster Genome Sequencing
I’d previously attempted to archive this data set on multiple occasions, across…
Manuscript Writing - Submitted!
2017
Genotype-by-sequencing at BGI
(http://eagle.fish.washington.edu/Arabidopsis/20170419_Overleaf_sci_data_oly_gbs_submission.png)
Manuscript - Oly GBS 14 Day Plan
2017
Genotype-by-sequencing at BGI
For Pub-a-thon 2017, Steven has asked us to put together a 14 day plan for our manuscripts.
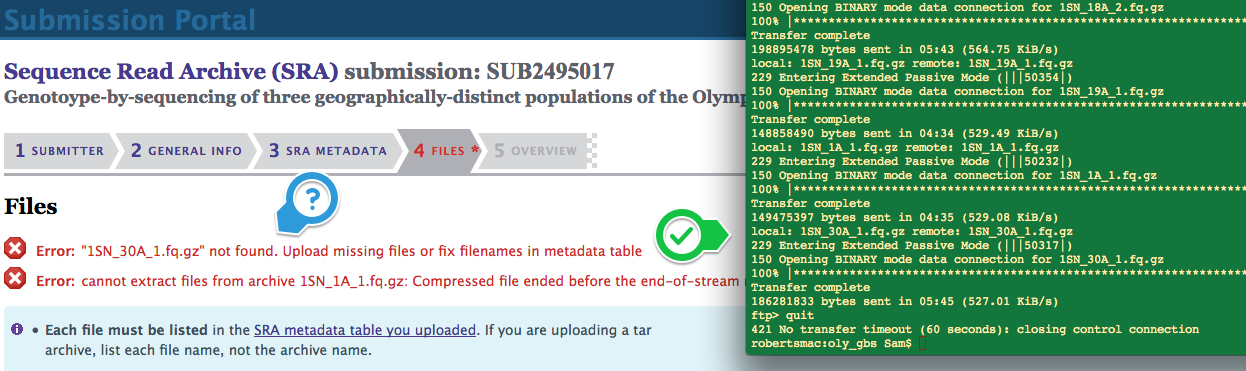
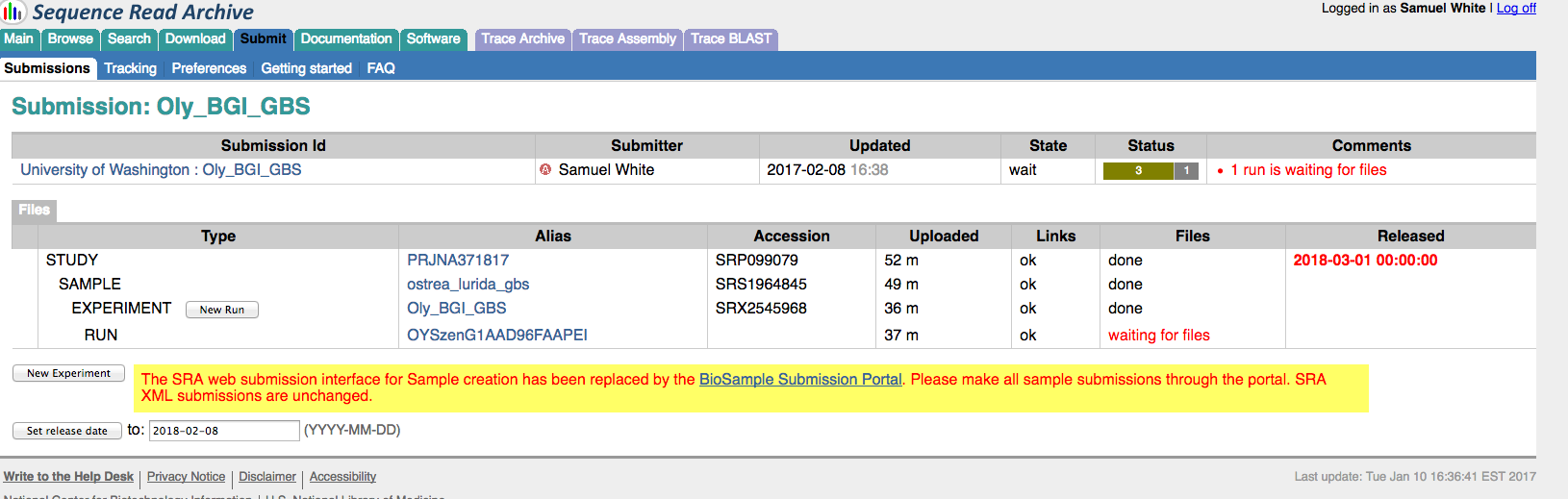
Data Management – SRA Submission Oly GBS Batch Submission
2017
Genotype-by-sequencing at BGI
An earlier attempt at submitting these files failed.
Data Management - SRA Submission Oly GBS Batch Submission Fail
2017
Genotype-by-sequencing at BGI
[I had previously submitted the two…
Computing - Oly BGI GBS Reproducibility Fail
2017
Genotype-by-sequencing at BGI
Since we’re preparing a manuscript that relies on BGI’s…
FASTQC - Oly BGI GBS Raw Illumina Data Demultiplexed
2017
Genotype-by-sequencing at BGI
Last week, I ran the two raw FASTQ files through FastQC. As expected, FastQC detected “errors”. These errors are due to the presence of adapter sequences, barcodes, and the use of a restriction enzyme (ApeKI) in library…
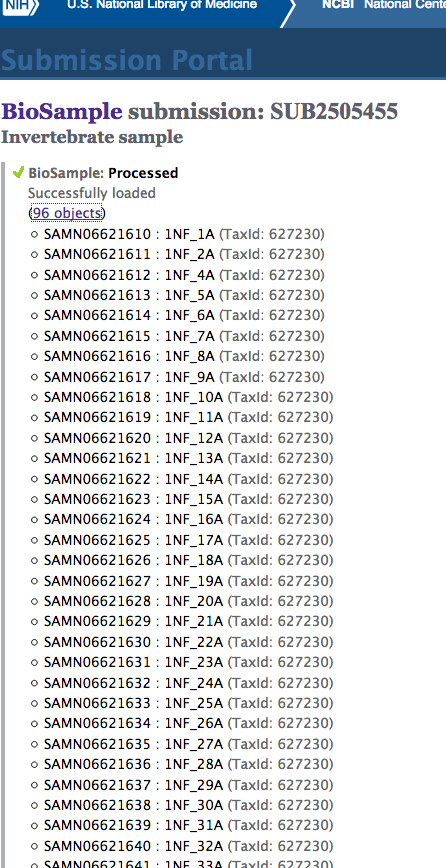
Data Management - SRA Submission of Ostrea lurida GBS FASTQ Files
2017
Genotype-by-sequencing at BGI
SRA Submissions
Prepared a short read archive (SRA) submission for archiving our Olympia oyster genotype-by-sequencing (GBS) data in NCBI. This is in preparation for submission of the mansucript we’re putting…
Sample Submission – Geoduck gDNA for Illumina Pilot Sequencing Project
2017
Geoduck Genome Sequencing
Samples Submitted
Sent 10μg of the geoduck gDNA I isolated earlier today to Illumina on dry ice via FedEx Standard Overnight service.
Data Management - Replacement of Corrupt BGI Oly Genome FASTQ Files
2017
Olympia Oyster Genome Sequencing
Previously, Sean and Steven identified two potentially corrupt FASTQ files. I contacted BGI about getting replacement files and they informed me that all versions of the FASTQ files they have delivered on three separate occasions are all the same…
Sample Submission - Geoduck Tissue & gDNA for Illumina Pilot Sequencing Project
2016
Geoduck Genome Sequencing
Samples Submitted
Sent the following samples to Illumina for possible selection in a new pilot sequencing platform they’re working on.
Sample Submission - Ostrea lurida gDNA for PacBio Sequencing
2016
Olympia Oyster Genome Sequencing
Samples Submitted
Submitted 10μg (30.7μL) of the O.lurida gDNA I isolated on 20161214 to the UW PacBio facility - Order #450.
Data Management - Integrity Check of Final BGI Olympia Oyster & Geoduck Data
2016
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
After completing the downloads of these files from BGI, I needed to verify that the downloaded copies matched the originals. Below is a Jupyter Notebook detailing how I…
Data Managment - Trim Output Cells from Jupyter Notebook
2016
Miscellaneous
Last week I downloaded the final BGI data files and assemblies for Olympia oyster and geoduck genome sequencing projects. However, the output from the download command made the Jupyter Notebook files too large to view and/or upload to GitHub. So, I had to trim the output cells…
Data Management - Download Final BGI Genome & Assembly Files
2016
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
We received info to download the final data and genome assembly files for geoduck and Olympia oyster from BGI.
Data Analysis - Continued O.lurida Fst Analysis from GBS Data
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Continued the analysis I started the other day. Still following Katherine Silliman’s notebook for guidance.
Data Analysis - Initial O.lurida Fst Determination from GBS Data
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Finally running some analysis on the output from my PyRad analysison 20160727.
Data Management - Tracking O.lurida FASTQ File Corruption
2016
Olympia Oyster Genome Sequencing
UPDATE 20170104 - These two corrupt files have been replaced with non-corrupt files.
Computing - Retrieve data from Amazon EC2 Instance
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
I had an existing instance that still had data on it from my PyRad analysis on 20160727 that I needed to retrieve.
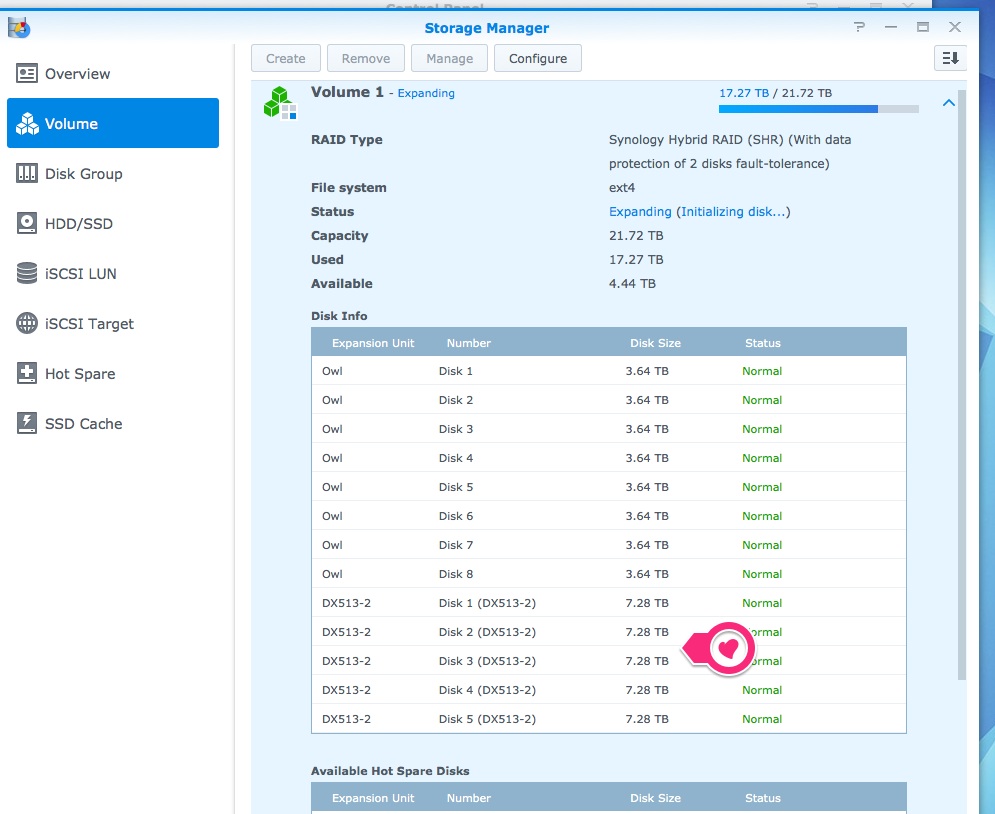
Server HDD Failure – Owl
2016
Computer Servicing
Noticed that Owl (Synology DS1812+ server) was beeping.
Computing - Amazon EC2 Instance Out of Space?
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Running PyRad analysis on the Olympia oyster GBS data. PyRad exited with warnings about running out of space. However, looking at free disk space on the EC2 Instance…
Data Analysis - Oly GBS Data Using Stacks 1.37
2016
Genotype-by-sequencing at BGI
This analysis ran (or, more properly, was attempted) for a couple of weeks and…
SRA Release - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck)
2016
Protein expression profiles during sexual maturation in Geoduck
SRA Submissions
The RNAseq data that [I previously submitted to NCBI short read archive…
Data Management - O.lurida Raw BGI GBS FASTQ Data
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
BGI had previously supplied us with demultiplexed GBS FASTQ files. However, they had not provided us with the information/data on how those files were created. I contacted…
Data Management - Concatenate FASTQ files from Oly MBDseq Project
2016
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Steven requested I concatenate the MBDseq files we received for this project:
Data Analysis - Oly GBS Data from BGI Using Stacks
2016
Genotype-by-sequencing at BGI
UPDATE (20160418) : I’m posting this more for posterity, as Stacks continually…
SRA Submission – Genome sequencing of the Olympia oyster (Ostrea lurida)
2016
Olympia Oyster Genome Sequencing
SRA Submissions
Adding our Olympia oyster genome sequencing (sequencing done by BGI) to the NCBI Sequence Read Archive (SRS). The current status can be seen in the screen cap below. Release…
SRA Submission - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck).
2016
Protein expression profiles during sexual maturation in Geoduck
RNAseq experiment, which is…
Data Management - O.lurida 2bRAD Dec2015 Undetermined FASTQ files
2016
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
An astute observation by Katherine Sillimanrevealed that the FASTQ files I had moved to our high-throughput sequencing archive on our server Owl, only had two FASTQ files labeled as “undetermined”. Based on the…
Data Received - Ostrea lurida MBD-enriched BS-seq
2016
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Received the Olympia oyster, MBD-enriched BS-seq sequencing files (50bp, single read) from ZymoResearch (submitted 20151208). Here’s the sample list:
Data Received - Ostrea lurida genome sequencing files from BGI
2016
Olympia Oyster Genome Sequencing
Downloaded data from the BGI project portal to our server…
Data Received - Panopea generosa genome sequencing files from BGI
2016
Geoduck Genome Sequencing
Downloaded data from the BGI project portal to our server…
Data Received - Bisulfite-treated Illumina Sequencing from Genewiz
2016
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Received notice the sequencing data was ready from Genewiz for the samples submitted 20151222.
Data Received - Oly 2bRAD Illumina Sequencing from Genewiz
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
The data was made available to use on 20151224 and took two days to download.
Sample Submission - BS-seq Library Pool to Genewiz
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Samples Submitted
Pooled 10ng of each of the libraries prepared yesterday with [TruSeq DNA Methylation Library Kit…
Illumina Methylation Library Quantification - BS-seq Oly/C.gigas Libraries
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Re-quantified the libraries that were completed yesterday using the…
Illumina Methylation Library Construction - Oly/C.gigas Bisulfite-treated DNA
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Took the bisulfite-treated DNA from 20151218 and made Illumina libraries using the [TruSeq DNA Methylation Library Kit…
Bioanalyzer - Bisulfite-treated Oly/C.gigas DNA
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Following the guidelines of the [TruSeq DNA Methylation Library Prep Guide…
Bisulfite Treatment - Oly Reciprocal Transplant DNA & C.gigas Lotterhos DNA for BS-seq
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
After confirming that the DNA available for this project looked good, I performed bisulfite treatment on the following gDNA samples:
Agarose Gel - Oly gDNA for BS-seq Libraries, Take Two
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
The gel I ran earlier today looked real rough, due to the fact that I didn’t bother to equalize…
Agarose Gel - Oly gDNA for BS-seq Libraries
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Ran 1μL of each sample from yesterday’s DNA isolation on a 0.8% agarose, low-TAE gel, stained with ethidium bromide.
DNA Isolation - Oly gDNA for BS-seq
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Need DNA to prep our own libraries for bisulfite-treated high-throughput sequencing (BS-seq).
Sample Submission - 2bRAD Libraries for Genewiz
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Samples Submitted
Pooled the libraries into a single sample for sequencing on Illumina HiSeq2500 by Genewiz.
Sample Submission - Olympia oyster MBD-enriched DNA to ZymoResearch
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Samples Submitted
We opted to go with ZymoResearch for this project because they could do the bisulfite treatment and finish the sequencing by the end of January.
Sample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI
2015
Olympia Oyster Genome Sequencing
Samples Submitted
Yep, BGI still needs more gDNA for the Olympia oyster genome sequencing…
Sample Submission - Oly Oyster Bay Tissues for GBS
2015
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Samples Submitted
Sent Olympia oyster tissue samples to BGI for genotype-by-sequencing (GBS). Tissues were shipped FedEx standard overnight on dry ice.
Samples Received - Oly Tissue & DNA from Katherine Silliman
2015
Olympia oyster reciprocal transplant
Samples Received
Samples were stored @ -20C in FTR 209.
DNA Quality Assessment - Geoduck & Olympia Oyster gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Have three separate sets of geoduck & olympia oyster gDNA that need to be run on gels before sending to BGI for genome sequencing:
Phenol-Chloroform DNA Cleanup - Geoduck gDNA
2015
Geoduck Genome Sequencing
The gDNA I extracted on 20151104 didn’t look great on the NanoDrop so I…
Phenol-Chloroform DNA Cleanup - Olympia Oyster gDNA
2015
Olympia Oyster Genome Sequencing
The gDNA I extracted on 20151104 didn’t look great on the NanoDrop so I…
DNA Quantification - MBD-enriched Olympia oyster DNA
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Quantified the MBD enriched samples prepped over the last two days: MBD enrichment, EtOH precipiation.
Ethanol Precipitation - Olympia oyster MBD
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Precipitated the MBD enriched DNA from yesterday according to the MethylMiner Methylated DNA Enrichment Kit (Invitrogen) protocol.
MBD Enrichment - Sonicated Olympia Oyster gDNA
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Olympia oyster gDNA that had previously been sonicated and fragmented …
DNA Sonication - Oly gDNA for MBD
2015
MBD Enrichment for Sequencing at ZymoResearch
In preparation for MBD enrichment, fragmented Olympia oyster gDNA with a target size of ~350bp.
qPCR – Oly RAD-Seq Library Quantification
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
After yesterday’s attempt at quantification revealed insufficient dilution of the libraries, I repeated the qPCRs using 1:100000 dilutions of each of the libraries. Used the KAPA Illumina Quantification Kit (KAPA Biosystems) according to the…
qPCR - Oly RAD-Seq Library Quantification
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
The final step before sequencing these 2bRAD libraries is to quantify them. …
Gel Extraction - Oly RAD-Seq Prep Scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Extracted the PCR products from the gel slices from 20151113 using the QIAQuick Gel Extraction Kit (Qiagen) according to the manufacturer’s protocol.…
DNA Quality Assessment - Geoduck, Oly & Oly 2SN
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Olympia oyster reciprocal transplant
I recently ran gDNA isolated for geoduck and Olympia oyster genome sequencing, as well as gDNA isolated from the Olympia oyster reciprocal transplant experiment out on a Bioanalyzer (Agilent) using the DNA 12000 chips. The results from the chip were a bit confusing and difficult to assess exactly what was going…
PCR – Oly RAD-seq Prep Scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continuing with the RAD-seq library prep. Following the Meyer Lab 2bRAD protocol. After…
PCR – Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continuing with the RAD-seq library prep. Following the Meyer Lab 2bRAD protocol.
DNA Quantification & Quality Assessment - Oly 2SN gDNA
2015
Olympia oyster reciprocal transplant
Comparison of three different approaches to using the E.Z.N.A. Mollusc Kit:
DNA Quantification & Quality Assessment - Geoduck & Oly gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Quantif…
DNA Isolations – Oly Fidalgo 2SN Ctenidia
2015
Olympia oyster reciprocal transplant
Isolated DNA from 24 2SN ctenidia samples from Friday’s sampling (#32 - 55). Samples were thawed at RT.
Oyster Sampling - Oly Fidalgo 2SN, 2HL, 2NF Reciprocal Transplants Final Samplings
2015
Olympia oyster reciprocal transplant
The remaining Olympia oysters from Jake…
DNA Isolation – Geoduck & Olympia Oyster
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Amazingly, we need more gDNA for the two genome sequencing projects (geoduck and Olympia oyster). Used geoduck adductor muscle sample from Box 1 of the geoduck samples collected by Brent &…
Adaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continued to follow the 2bRAD protocol (PDF) developed by Eli Meyer’s lab.
Restriction Digest – Oly gDNA for RAD-seq w/AlfI
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
The previous attempt at making these RAD libraries failed during the prep-scale PCR, likely due to a discrepancy in the version of the Meyer Lab protocol I was following, so I have to start at the beginning to try to make these…
Troubleshooting - Oly RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Oddly, it turns out that…
DNA Isolations - Fidalgo 2SN Reciprocal Transplants Final Samplings
2015
Olympia oyster reciprocal transplant
The remaining Olympia oysters from Jake…
PCR - Oly RAD-seq Prep Scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continuing with the RAD-seq library prep. Following the Meyer Lab 2bRAD protocol.
PCR - Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continuing with the RAD-seq library prep. Following the Meyer Lab 2bRAD protocol.
Adaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continued to follow the 2bRAD protocol (PDF) developed by Eli Meyer’s lab.
Restriction Digest – Oly gDNA for RAD-seq w/AlfI
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Used a subset (10 samples) from the Ostrea lurida gDNA isolated 20150916 to prepare RAD libraries.
Sample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI
2015
Olympia Oyster Genome Sequencing
Samples Submitted
Previous shipment of gDNA proved to be of insufficient quantity when assessed by BGI, so needed to isolate more.
Agarose Gel - Geoduck & Olympia Oyster gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Olympia oyster reciprocal transplant
Needed to assess the integrity of the newest gDNA isolated for the two genome sequencing projects: Geoduck gDNA from earlier today and Olympia oyster gDNA from 20151002.
DNA Quantification - Pooled geoduck gDNA
2015
Geoduck Genome Sequencing
Pooled the gDNA samples from earlier today & from 20151002.
DNA Isolation - Geoduck & Olympia Oyster
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Olympia oyster reciprocal transplant
Amazingly, we need more gDNA for the two genome sequencing projects (geoduck and Olympia oyster). Used geoduck “foot 1” sample from Box 1 of the foot samples…
PCR - Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Yesterday’s test scale PCR failed to produce any bands in any samples (expected size of ~166bp). This is not particularly surprising, due to the…
PCR - Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Reagent Prep
Continuing with the RAD-seq library prep. Following the Meyer Lab 2bRAD protocol.
Adaptor Ligation - Oly AlfI-Digested gDNA for RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Reagent Prep
Yesterday’s AlfI over night restriction digest was heat inactivated by heating @ 65C for 10mins. Samples were stored on ice.
Restriction Digest - Oly gDNA for RAD-seq w/AlfI
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Reagent Prep
Used a subset (10 samples) from the Ostrea lurida gDNA isolated 20150916 to prepare RAD libraries. This will be done to assess whether or not these samples, which appear to…
Sample Submission - Olympia Oyster gDNA for Genome Sequencing @ BGI
2015
Olympia Oyster Genome Sequencing
Samples Submitted
Shipped the pooled gDNA we’ve been accumulating to BGI to initiate the Olympia oyster genome sequencing project.
Agarose Gel - Geoduck & Olympia oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Ran a 0.8% agarose, 1x modified TAE gel (w/EtBr) with geoduck and Olympia oyster gDNA that was precipitated earlier today. Used 5μL of each sample (~500ng).
Ethanol Precipitation - Geoduck & Olympia oyster gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Pooled all of the geoduck gDNA from all the previous geoduck isolations for the Geoduck Genome Sequencing Project and pooled all of the Ostrea lurida gDNA from previous…
Agarose Gel - Olympia oyster Whole Body gDNA Integrity Check
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Ran the gDNA isolated yesterday from Ostrea lurida whole body on a 0.8% modified TAE gel (w/EtBr) to assess gDNA integrity. Used 1μL of each sample.
DNA Isolation - Olympia oyster whole body
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Continued the extractions that Steven began yesterday and this morning using the E.Z.N.A. Mollusc DNA Kit (Omega Bio-Tek) after the RNase treatment @ 70C.
Agarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Ran 0.8% agarose 1x modified TAE gel stained with EtBr to assess the integrity of geoduck gDNA and Olympia oyster gDNA isolated earlier today.
Agarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Ran 0.8% agarose 1x modified TAE gel stained with EtBr to assess the integrity of geoduck gDNA and Olympia oyster gDNA isolated yesterday.
Genomic DNA Isolation - Olympia oyster adductor musle & mantle
2015
Olympia Oyster Genome Sequencing
Previously isolated gDNA from these tissues on 20150901. However, found out after the…
Agarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Ran…
Server Email Notifications Fix - Eagle
2015
Computer Servicing
The system was previously set to use Steven’s Comcast SMTP…
RAD-Seq Library Prep Reagents
2015
2bRAD Library Tests for Sequencing at Genewiz
Reagent Prep
A box with the above title was established in the -20C in FTR 209 containing the following:
Reverse Transcription – O.lurida DNased RNA 1hr post-mechanical stress
2015
Olympia oyster reciprocal transplant
Performed reverse transcription on the Olympia oyster DNased RNA from the 1hr post-mechanical stress samples from Jake’s project. To accommodate the large numbers of anticipated genes to be targeted in subsequent qPCRs, I prepared 100μL reactions (normally, 25μL reactions are…
RNA Quantification - O.lurida 1hr post-mechanical heat stress DNased RNA
2015
Olympia oyster reciprocal transplant
DNased RNA from 07272015 was quantified using the Roberts Lab NanoDrop1000.
RNA Isolation – O.lurida Ctenidia 1hr Post-Mechanical Stress
2015
Olympia oyster reciprocal transplant
Isolated RNA from [Jake’s Ostrea lurida ctenidia samples…
RNA Isolation - O.lurida Ctenidia 1hr Post-Mechanical Stress
2015
Olympia oyster reciprocal transplant
Isolated RNA from [Jake’s Ostrea lurida ctenidia samples…
Automatic Notebook Backups - wget Script & Synology Task Scheduler
2015
Computer Servicing
I’ve been tweaking a [shell script (notebook_backups.sh)(https://github.com/sr320/LabDocs/blob/master/code/script-box/notebook_backups.sh) to use the shell program
wget to retrieve fully functional HTML versions of our online notebooks for offline viewing. I had been planning on…RNAseq Data Receipt - Geoduck Gonad RNA 100bp PE Illumina
2015
Protein expression profiles during sexual maturation in Geoduck
Received notification that the samples sent on 20150601 for RNAseq were completed.
Reverse Transcription - O.lurida DNased RNA Controls and 1hr Heat Shock
2015
Miscellaneous
Performed reverse transcription on the Olympia oyster DNased RNA from the control samples and the 1hr heat shock samples of Jake’s project. To accommodate the large numbers of anticipated genes to be targeted in subsequent qPCRs, I prepared 100μL reactions (normally, 25μL reactions are…
Sample Submission – Olympia oyster & Sea Pen PCRs Sanger Sequencing
2015
Miscellaneous
Prepared two DNA plates and corresponding primer plates for sequencing at the UW HTGC from the purified gel-purified PCRs from yesterday. Primer plates were prepared by adding 7μL of NanoPure H2O to each well and then adding 3μL of 10μM primer to the appropriate wells. For the DNA plates…
Gel Purification - Olympia Oyster and Sea Pen PCRs
2015
Miscellaneous
Purified DNA from the remaining PCR bands excised by Jake on 20150609 and 20150610, as well as Jonathan’s sea pen PCRs from 20150604, using Ultrafree-DA spin columns (Millipore). Transferred gel pieces from storage tubes (1.5mL snap cap tubes) to spin columns. Spun 10,000g, 5mins @ RT. Transferred…
Sample Submission - Geoduck Gonad for RNA-seq
2015
Protein expression profiles during sexual maturation in Geoduck
Prepared two pools of geoduck RNA for RNA-seq (Illumina HiSeq2500, 100bp, PE) with GENEWIZ, Inc.
Bioanalyzer - Geoduck Gonad RNA Quality Assessment
2015
Protein expression profiles during sexual maturation in Geoduck
Before proceeding with transcriptomics for this project, we need to assess the integrity of the RNA via Bioanalyzer.
Reverse Transcription - Subset of Jake’s O.lurida DNased RNA
2015
Miscellaneous
Currently don’t have sufficient reagents to perform reverse transcription on the entire set of DNased RNA (control O.lurida ctenidia samples). To enable Jake to start testing out some of his primers while we wait for reagents to come in…
Bioinformatics – Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Previously trimmed the first 39 bases of sequence from reads from the BS-Seq data in an attempt to improve our ability to map the reads back to the C.gigas genome. However, Mac (and Steven) noticed that the last ~10 bases of all the reads showed a steady increase in the %G, suggesting some sort of…
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
UPDATE 20150528: The RNA isolated in this notebook entry may have been consolidated on 20150528.
Bioinformatics - Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
In…
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
UPDATE 20150528: The RNA isolated in this notebook entry may have been consolidated on 20150528.
BLAST - C.gigas Larvae OA Illumina Data Against GenBank nt DB
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
In an attempt to figure out what’s going on with the Illumina data we recently received for these samples, I BLASTed the 400ppm data set that had previously been de-novo assembled by…
Goals - May 2015
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Goals
Lineage-specific DNA methylation patterns in developing oysters
LSU C.virginica Oil Spill MBD BS Sequencing
Protein expression profiles during sexual maturation in Geoduck
Here are the things I plan to tackle throughout the month of May:
BLASTN - C.gigas OA Larvae to C.gigas Ensembl 1.24 BLAST DB
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
In an attempt to figure out what’s going on with the Illumina data we recently received for these samples, I BLASTed the 400ppm data set that had previously been de-novo assembled by…
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
UPDATE 20150528: The RNA isolated in this notebook entry may have been consolidated on 20150528.
Bioanalyzer Data - Geoduck RNA from Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
I received the Bioanalyzer data back for the geoduck foot RNA samples I submitted 20150422. The two samples were run on the RNA Pico chip assay.
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
UPDATE 20150528: The RNA isolated in this notebook entry may have been consolidated on 20150528.
Bioanalyzer Submission - Geoduck Gonad RNA from Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Submitted 3μL (~75ng) of RNA from each of the two gonad samples isolated from foot tissue embedded in paraffin histology blocks 20150408 (to…
Quality Trimming - C.gigas Larvae OA BS-Seq Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Jupyter (IPython) Notebook: 20150414_C_gigas_Larvae_OA_Trimmomatic_FASTQC.ipynb
Quality Trimming - LSU C.virginica Oil Spill MBD BS-Seq Data
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Jupyter (IPython) Notebook: 20150414_C_virginica_LSU_Oil_Spill_Trimmomatic_FASTQC.ipynb
Sequence Data Analysis - LSU C.virginica Oil Spill MBD BS-Seq Data
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Performed some rudimentary data analysis on the new, demultiplexed data downloaded earlier today:
Sequence Data Analysis - C.gigas Larvae OA BS-Seq Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Compared total amount of data generated from each index. The commands below send the output of…
Sequence Data - C.gigas OA Larvae BS-Seq Demultiplexed
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
I had previously contacted Doug Turnbull at the Univ. of Oregon Genomics Core Facility for help demultiplexing this data, as it was…
Sequence Data - LSU C.virginica Oil Spill MBD BS-Seq Demultiplexed
2015
LSU C.virginica Oil Spill MBD BS Sequencing
I had previously contacted Doug Turnbull at the Univ. of Oregon Genomics Core Facility for help demultiplexing this data, as it was…
RNA Isolation - Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Isolated RNA from geoduck gonad previously preserved with the PAXgene Tissue Fixative and Stabilizer and then embedded in paraffin blocks. See Grace’s notebook for…
Sequencing Data - C.gigas Larvae OA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Our sequencing data (Illumina HiSeq2500, 100SE) for this project has completed by Univ. of Oregon Genomics Core Facility (order number 2212).
TruSeq Adaptor Counts – LSU C.virginica Oil Spill Sequences
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Initial analysis, comparing barcode identification methods, revealed the following info about demultiplexing on untrimmed sequences:
TruSeq Adaptor Identification Method Comparison - LSU C.virginica Oil Spill Sequences
2015
LSU C.virginica Oil Spill MBD BS Sequencing
We recently received Illumina HiSeq2500 data back from this project. Initially looking at the data, something seems off. Using FASTQC, the quality drops of drastically towards the last 20…
DNA Quantification - Claire’s C.gigas Sheared DNA
2015
Lineage-specific DNA methylation patterns in developing oysters
In an attempt to obtain the most accurate measurement of Claire’s sheared, heat shock mantle DNA, I quantified the samples using a third method: fluorescence.
Library Quality Assessment - C.gigas OA larvae Illumina libraries
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Ran the 400ppm library preps on a DNA1000 Assay Chip (Agilent) on the Agilent 2100 Bioanalyzer.
BS-seq Library Prep - C.gigas Larvae OA 1000ppm
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Pooled 200ng each of the sheared 1B1 (4μL) & 1B2 (used the entire sample, 20μL) 5.13.11 1000ppm C.gigas larvae DNA samples for a total of 400ng. Total volume = 24μL.
DNA Quantification - C.gigas Larvae 1000ppm
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
After the discovery that there…
DNA Quantification - Claire’s Sheared C.gigas Mantle Heat Shock Samples
2015
Lineage-specific DNA methylation patterns in developing oysters
I previously checked Claire’s sheared DNA on the Bioanalyzer to verify the fragment size and to quantify the samples. Looking at her notebook, her numbers differ greatly from the Bioanalyzer, possibly due to the fact that the DNA1000 assay chip used only measures DNA fragments up to 1000bp in size. If her…
Bioanalyzer - C.gigas Sheared DNA from 20140108
2015
Lineage-specific DNA methylation patterns in developing oysters
To complement MBD ChiP-seq data and RNA-seq data that we have from this experiment, we want to generate, at a minimum, some BS-seq data from the same C.gigas individuals used for the other aspects…
Library Prep - Quantification of C.gigas larvae OA 1000ppm library
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
The completed BS Illumina library made on Friday (1000ppm) was quantified via fluorescence using the Quant-iT DNA BR Kit (Life Technologies/Invitrogen). Also quantified Jake’s…
Sequencing Data - LSU C.virginica MBD BS-Seq
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Our sequencing data (Illumina HiSeq2500, 100SE) for this project has completed by Univ. of Oregon Genomics Core Facility (order number 2112).
Bisulfite NGS Library Prep - Bisulfite Conversion & Illumina Library Construction of C.gigas larvae DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
The previous attempt at constructing a library for the 1000ppm larvae samples failed. I had previously sheared, quantified, and concentrated the DNA from this sample. As I had done previously, I combined 50ng from each of the two 1000ppm samples for a total of 100ng, and brought the sample…
Bisuflite NGS Library Prep – C.gigas larvae OA bisulfite library quantification
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
The two completed BS Illumina libraries (400ppm and 1000ppm) were quantified via fluorescence using the Quant-iT DNA BR Kit (Life Technologies/Invitrogen). Used 1uL of …
Bisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA (continued from yesterday)
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Cont…
Bisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
The two pooled bisulfite-treated DNA samples (400ppm and 1000ppm) from 20150114 were used to prepare bisulfite Illumina libraries with [EpiNext Post-Bisulfite DNA Library Preparation Kit (Illumina)…
Library Cleanup - LSU C.virginica MBD BS Library
2015
LSU C.virginica Oil Spill MBD BS Sequencing
I was contacted by the sequencing facility at the…
DNA Bisulfite Conversion - C.gigas larvae OA Sheared DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
After discussing with Steven, decided to use only samples from 20110513, due to high algae amounts present in the 20110519 samples.
SpeedVac - C.gigas larvae OA DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
The DNA extracted and sheared on 20150109 was in a volume too great to proceed with bisulfite conversion. Dried…
DNA Isolation - C.gigas larvae from 2011 NOAA OA Experiment
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Some tubes contained a high quantity of algae, based on quantity of material in tube and overall green color.
Bisulfite NGS Library Prep - LSU C.virginica Oil Spill MBD Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA (continued from yesterday)
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
LSU C.virginica Oil Spill MBD BS Sequencing
Continued library prep from yesterday. Set up Library Amplification according to the protocol. The samples received the following Barcode Indices:
Bisulfite NGS Library Prep - LSU C.virginica Oil Spill Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
LSU C.virginica Oil Spill MBD BS Sequencing
Constructed next generation libraries (Illumina) using the bisulfite-treated DNA from yesterday using the EpiNext Post-Bisulfite DNA…
Bisulfite Conversion - LSU C.virginica Oil Spill MBD DNA and Emma’s C.gigas Larvae OA DNA
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
LSU C.virginica Oil Spill MBD BS Sequencing
Performed bisulfite conversion on MBD DNA samples from LSU C.virginica oil spill samples (see 201411202 and 20141126) and Emma’s C.gigas larvae OA DNA samples (see…
DNA Isolation - Claire’s C.gigas Female Gonad for Illumina Bisulfite Sequencing
2014
Lineage-specific DNA methylation patterns in developing oysters
Due to poor “tag counts” from the initial sequencing (DATE) and the re-sequencing (20131127) of this sample, the HTGU facility has concluded that…
EtOH Precipitation - LSU C.virginica Oil Spill MBD Continued (from 20141126)
2014
LSU C.virginica Oil Spill MBD BS Sequencing
Precipitation was continued according to the MethylMiner Methylated DNA Enrichment Kit…
Gel - Sheared gDNA
2014
LSU C.virginica Oil Spill MBD BS Sequencing
Ran ~250ng (out of 3000ng, according to Claire) of LSU C.virginica oil spill gDNA on a gel that was previously sheared by Claire to verify that shearing was successful.
DNA Isolation - C.gigas Larvae from Emma OA Experiments
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Isolated gDNA using DNazol from the following larval samples for potential MBD selection and bisulfite sequencing:
RAD Sequencing - Oly Oyster gDNA-01 RAD Library (from 20141110)
2014
Olympia oyster reciprocal transplant
Samples Submitted
Prepared 10nM of the library in total volume of 20uL of Buffer EB (Qiagen) + 0.1% Tween-20, according the University of Oregon’s Genomic’s Core:
Library Prep - Oly Oyster gDNA-01 RAD
2014
Olympia oyster reciprocal transplant
Used gel-purified, size-selected DNA from yesterday to prepare the RAD library using the Kappa LTP Kit:
Ligation - Illumina P1 Adapters for Oly Oyster gDNA-01 RAD Sequencing (from 20141031)
2014
Olympia oyster reciprocal transplant
Made 25nM working stock from the 100nM stock…
Restriction Digest - Oly Oyster gDNA-01 for RAD Sequencing (from 20141029)
2014
Olympia oyster reciprocal transplant
Samples required two days of drying for all samples to fully dry down.
DNA Allocation - Oly Oyster gDNA-01 for RAD Sequencing (from 20141022)
2014
Olympia oyster reciprocal transplant
Transferred 500ng of…
Package - Received Package from Jerome LaPeyre from LSU
2014
LSU C.virginica Oil Spill MBD BS Sequencing
Oyster (C.virginica)…
DNA Isolation - Claire’s C.gigas Female Gonad
2014
Lineage-specific DNA methylation patterns in developing oysters
Trying this sample again(!!), but will now use TE for pellet resuspension to prevent sample degradation. Incubated sample RT on rotator in 500uL of DNazol + 2.7uL of…
DNA Isolation - Test Sample
2014
Lineage-specific DNA methylation patterns in developing oysters
Miscellaneous
Due to the recent poor quality gDNA that has been isolated from C.gigas gonad, I decided to do a quick test…
Phenol-Chloroform DNA Clean Up - Mac and Claire’s Samples (from 20140410)
2014
Lineage-specific DNA methylation patterns in developing oysters
Due to low 260/230 values and Mac’s smeary sample, performed a phenol-chloroform DNA cleanup on the samples isolated 20140410.
DNA Isolation - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad
2014
Lineage-specific DNA methylation patterns in developing oysters
Due to the…
Cloned Hard Drive - Windows XP Opticon Computer (Aquacul8)
2014
Computer Servicing
Cloned the XP hard drive…
DNA gel - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad
2014
Lineage-specific DNA methylation patterns in developing oysters
Ran out 2uL of Clair’es C.gigas female gonad gDNA (from 20140328) and Mac’s…
DNA Isolation - C.gigas Female Gonads (from frozen)
2014
Lineage-specific DNA methylation patterns in developing oysters
Isolated gDNA from Claire’s “Female DNA” (from 05/16/2013) using the Qiagen Blood & Tissue DNeasy Kit according to the manufacturer’s protocol, with the following changes:
Reverse Transcription - Herring RNA from 20091026
2013
Miscellaneous
Performed an RT reaction on pooled herring gonad and liver mRNA from 20091026 for James Raymond at the UNLV. A single RT reaction was performed using 12.75uL (208ng) of the pooled gonad mRNA and 5uL (132.5ng) of the pooled liver RNA, according to our…
Sample Submission - QPX RNA and DNA for Illumina 36bp single-end RNA/DNAseq
2012
Miscellaneous
Samples Submitted
Submitted samples to HTGU for Illumina sequencing.
qPCR - Detection of V.tubiashii Presence and Expression Using VtpA Primers in DNA/cDNA from yesterday
2012
Miscellaneous
Ran qPCR with VtpA primers on cDNA and DNA (from yesterday) of C.gigas larvae to see levels of V.tubiashii compared to their water filter samples (see 20120326). Master mix calcs are here. Plate layout, cycling params, etc can be seen in the qPCR Report (see Results). Used 1uL of cDNA and 100ng (1uL)…
qPCR - DNased RNA from earlier today
2012
Miscellaneous
Checked DNased RNA samples from earlier today for the presence of residual gDNA. Used C.gigas BB16 gDNA (from 20110201) diluted to ~7ng/uL as a positive control to match the dilution factor of the RNA that will be used in the reverse transcription reaction…
qPCR - C.gigas actin and GAPDH on V.vulnificus exposure cDNA (from 20110311)
2011
Miscellaneous
Ran a qPCR on all cDNA samples from the V.vulnificus exposure experiment from 20110111. This qPCR was to test 2 of 4 potential normalizing genes to evaluate which genes show the least amount of effect from the treatments in this experiment. Primers for actin…
qPCR - C.gigas 18s and EF1a on V.vulnificus exposure cDNA (from 20110311)
2011
Miscellaneous
Ran a qPCR on all cDNA samples from the V.vulnificus exposure experiment from 20110111. This qPCR was to test 2 of 4 potential normalizing genes to evaluate which genes show the least amount of effect from the treatments in this experiment. Primers for 18s…
Restriction Digestions - HpaII and MspI on Mac’s C.gigas gDNA Samples: Round 1
2010
Miscellaneous
Set up restriction digests for subsequent analysis by methylation specific PCR (MSP). This will be the first of two rounds of digestion with the same enzyme on each sample. Samples and master mixes are…
Package - Hard Clam Samples from Rutgers
2010
Miscellaneous
Rec’d package of hard clam samples from…
gDNA Isolation - Various gigas samples
2010
Miscellaneous
Placed ~20mg fragments of tissue in 250uL…
RNA Precipitation & mRNA Isolation for SOLiD Libraries - Pooled abalone total RNA: Carmel control, Carmel exposed
2010
Miscellaneous
RNA of 8 samples from each group was pooled equally from each individual. RNA was precipitated according to Ambion’s MicroPolyA Purist Kit. Used 0.1 volumes of 3M NaAOc…
PCR - Sepia cDNA (from yesterday)
2009
Miscellaneous
Set up PCR on recent Sepia cDNA samples using both S. officianalis_rhodopsin_F, R primers & Sep_op_F2, R2 primers. PCR set up is here. Looking back at my old paper (gasp!) notebook from March 2007, I noticed that each primer set required differing amounts of Mg2+. Rhodopsin primers require 3mM Mg2+ in the…
mRNA Isolation - Herring gonad/ovary RNA (from 20091023)
2009
Miscellaneous
Sample was spun 16,000g…
RNA Precipitation - Herring Liver Samples
2009
Miscellaneous
A subset of samples (4 samples from each group) were pooled (see spreadsheet, green-highlighted samples), each providing ~68ug of RNA. The pooled sample was split into two tubes (435uL/tube). 0.1 vols (43.5uL) of 3M NaOAC (pH=5.2) were added, then 2…
Spec Reading - C.pugetti gDNA from 20090526
2009
Miscellaneous
A recent email from JGI indicates that they are satisfied with the quality of DNA (as seen on 20090601), however their estimate of the gDNA concentration (42ng/uL) means that we have ~16ug of DNA. They requested 50ug. Based on the gel, their calculations are reasonable.…
DNA Methylation Test - Gigas site gDNA (BB & DH) from 20090515
2009
Miscellaneous
Used BB & DH samples #11-17 for procedure. Followed Epigentek’s protocol. My calcs for dilutions/solutions needed are here. All solutions were made fresh before using, except for Diluted GU1 which was made at the beginning of the procedure and stored on ice in a 50mL conical wrapped in aluminum…
DNase Treatment - Oyster RNA from yesterday
2009
PROPS
Yesterday’s qPCR indicated that all of the RNA still contained gDNA contamination. So, took 10ug of RNA from BB #1-10 and DH#1-10 (calcs/workup BB and DH) and brought the volumes up to 50uL with 0.1% DEPC-H2O. Processed the samples according to Ambion Tubrbo DNA-free protocol. Will…
PCR - Bay/Sea scallop gDNA isolated earlier today
2009
Miscellaneous
Used 3 sets of reverse primers:
cDNA - Abalone RNA from 20090331 & 20090402
2009
Miscellaneous
cDNA was made from the above RNA samples using the Qiagen Quantitect RT Kit. The samples were laid out in a PCR plate. 274.2ng of RNA was used in the rxn for each sample, based on the lowest concentration RNA sample (08:3-20) to equalize all the samples. The Genomic Wipeout step of the kit…
Sequencing - Y2H colony PCRs from 20081112
2008
Myostatin Interacting Proteins
Purified bands were delivered for sequencing. See sequencing log for plate layout/primers.
No matching items