Ran the gDNA isolated yesterday from Ostrea lurida whole body on a 0.8% modified TAE gel (w/EtBr) to assess gDNA integrity. Used 1μL of each sample.
Results:
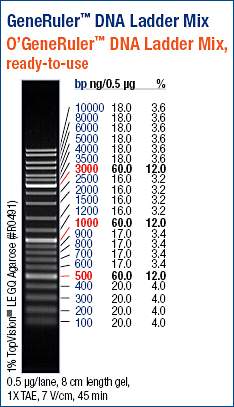 (https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
(https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)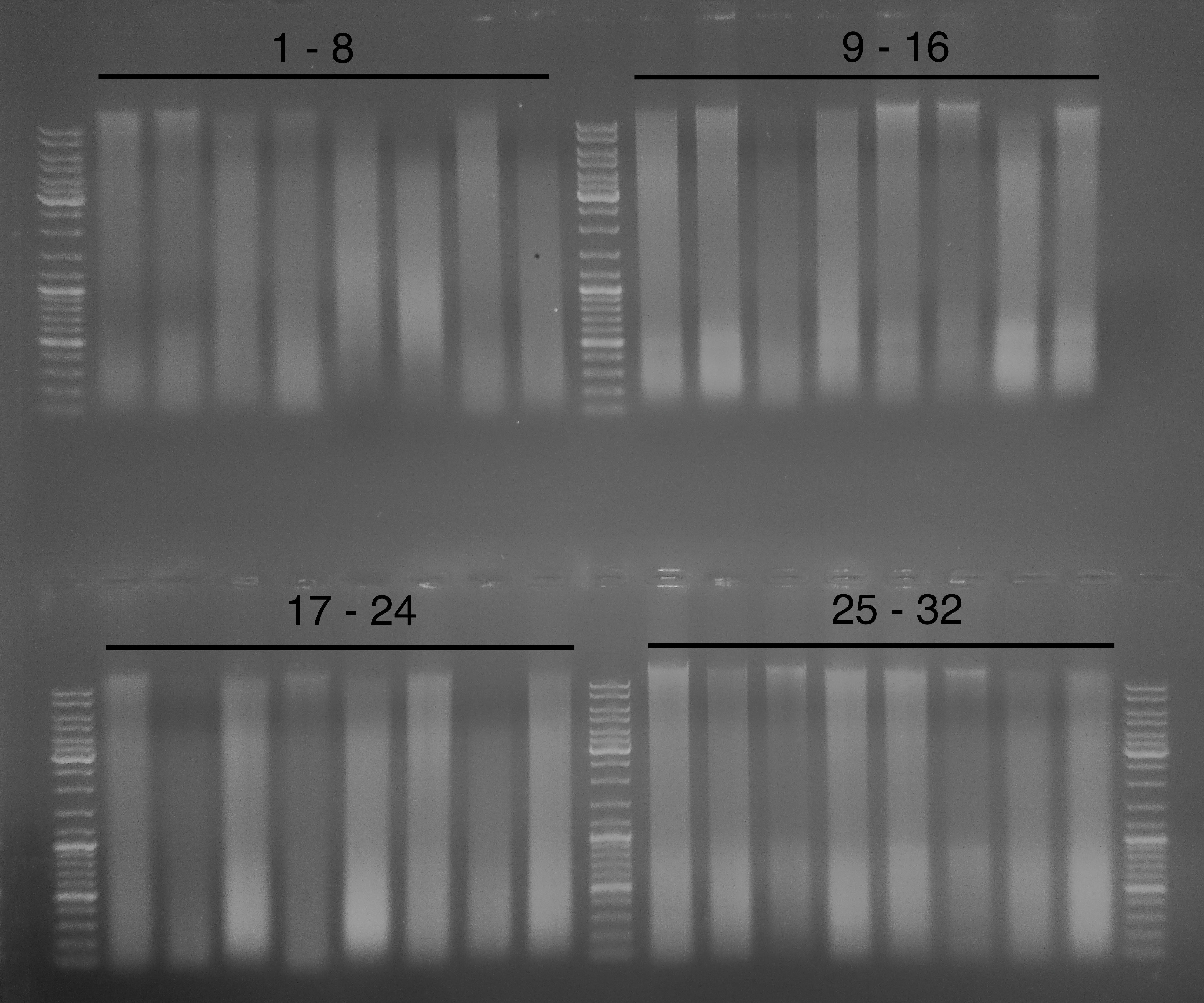
The results are not good. Every sample exhibits serious degradation (the smearing that’s present in each lane). There should be a distinct, high molecular weight band with no smearing if the gDNA was high quality and intact. These extractions also served as a comparison in slight differences in the extraction procedure (homogenization with & without mortar/pestle), as described in Steven’s post. However, those differences seem to have no impact on the quality of the resulting gDNA.
I [isolated gDNA from Ostrea lurida tissue samples two weeks ago using the E.Z.N.A. Mollusc DNA Kit (Omega Bio-Tek)(2015/09/01/agarose-gel-geoduck-olympia-oyster-gdna-integrity-check.html) and didn’t see this level of degradation. Additionally, Katherine Silliman used the E.Z.N.A. Mollusc DNA Kit to isolate gDNA from Ostrea lurida larvae and obtained high quality gDNA in virtually all of her samples. Below is my gel and Katherine’s gel for quick comparison to the one above:
[caption id=“” align=“alignleft” width=“301”] (http://eagle.fish.washington.edu/Arabidopsis/20150901_gDNA_geoduck_oly.jpg) Ostrea lurida gDNA isolated from adductor muscle & mantle tissues (lanes 4 & 5). Despite low quantity loading, notice that smearing below high molecular weight bands is limited to a low molecular weight range.[/caption]
(http://eagle.fish.washington.edu/Arabidopsis/20150901_gDNA_geoduck_oly.jpg) Ostrea lurida gDNA isolated from adductor muscle & mantle tissues (lanes 4 & 5). Despite low quantity loading, notice that smearing below high molecular weight bands is limited to a low molecular weight range.[/caption]
[caption id=“” align=“alignleft” width=“213”] (https://marinegenes.files.wordpress.com/2015/09/gel_9_11_15.jpg?w=584) Katherine’s gel of Ostrea lurida gDNA isolated from larvae.[/caption]
(https://marinegenes.files.wordpress.com/2015/09/gel_9_11_15.jpg?w=584) Katherine’s gel of Ostrea lurida gDNA isolated from larvae.[/caption]
I can’t be certain what is causing this issue. We previously had this same issue with a different group of Ostrea lurida whole body gDNA isolations (using a DNeasy Blood & Tissue Kit [Qiagen]). Two different kits using whole bodies and both sets of extractions have produced similarly bad results. It’s certainly possible that some nastiness (that’s a scientific term, btw) is being introduced by using whole body instead of specific tissues.
Another possible contributor to the DNA degradation we’ve seen is how the samples were collected and stored. I’m not up-to-date on exactly how the preservation was accomplished, but I do know that the Ostrea lurida whole body samples I previously worked with were just masses of black when I removed them from shells/tubes for isolation. So, in that case, it wasn’t terribly surprising that that the gDNA obtained from those was degraded. It should also be noted that Katherine’s extraction were from whole larvae that had been stored in RNAlater. Although a direct comparison cannot be made due to the difference in developmental stage between Katherine’s samples and these, it lends some evidence to the possibility that sample collection/storage is a contributor to the degraded gDNA we’re obtaining from whole body oyster extractions. However, with that being said, I’m not sure what the collection and storage background is on this particular set of samples.