---
title: "Designing Tunicate Primers"
author: "Celeste Valdivia"
date: "2023-10-18"
categories: [protocol, qPCR, primers, nickel chloride]
image: "bot_genome_proj.jpeg"
---
```{r setup, include=FALSE}
library(knitr)
library(tidyverse)
knitr::opts_chunk$set(echo = TRUE)
```
# Overview
I am interested in designing primers for *Botryllus schlosseri* to evaluate differences in expression of apoptosis, stemness, oxidative stress, and senescence related genes after an acute 24 hour exposure to two concentrations of nickel chloride, 5 mg/mL and 45 mg/mL, and a control.
# Potential Gene List
This list serves as a jumping off point for myself to look into genes that may potentially be differentially expressed in *Botryllus schlosseri* exposed to the clastogen (by oxidative damage to the DNA), nickel chloride. Will not be intending to use all of these genes but simply to look into some general areas to narrow down what genes would be most interesting and relevant to evaluate in my target organism.
| Role | Gene ID | Whole Gene Name |
|:---------------|:---------------|:--------------------------------------|
| Senescence | TP53 (p53) | Tumor protein p53 |
| Senescence | CDKN1A (p21) | Cyclin-dependent kinase inhibitor 1A |
| Senescence | CDKN2A (p16) | Cyclin-dependent kinase inhibitor 2A |
| Senescence | SIRT1 | Sirtuin 1 |
| Senescence | SIRT6 | Sirtuin 6 |
| Senescence | FOXO3 | Forkhead box O3 |
| Senescence | IGFBP3 | Insulin-like growth factor-binding protein 3 |
| Apoptosis | CASP3 | Caspase 3 |
| Apoptosis | CASP8 | Caspase 8 |
| Apoptosis | BCL2 | B-cell lymphoma 2 |
| Apoptosis | BAX | BCL2-associated X protein |
| Apoptosis | FAS | Fas cell surface death receptor (CD95) |
| Apoptosis | TNF | Tumor necrosis factor |
| Oxidative Stress | HSP70 | Heat Shock Protein 70 |
| Oxidative Stress | SOD | Superoxide Dismutase.
Various isoforms, such as SOD1, SOD2, and SOD3 |
| Oxidative Stress | CAT | Catalase enzyme |
| Oxidative Stress | HMOX1 | Heme oxygenase enzyme 1 |
| Oxidative Stress | NQO1 | NADPH quinone dehydrogenase 1 |
| stemness | MYC | Myc proto-oncogene protein |
| stemness | SOXB1 | SRY-Box Transcription Factor B1 |
| stemness | POU3 | POU Class 3 |
| aging and proliferation | HSPA9 | Mortalin |
# Two Approaches for Acquiring Primers
## Extensive search in the literature for pre-designed *Botryllus* specific primers.
For the purpose of expedience and accuracy. This generally is the preferred approach.
| Gene | NCBI Accession Number | DOI | Fwd Primer | Rev Primer |
|---------------|---------------|---------------|---------------|---------------|
| Mortalin | KY212120 | doi:10.1016/
j.ydbio.2017.10.023 | TTCATGATACCGAA
ACCAAGATGGACG | AATTTCCTCCTTG
ATGGCATCCACC |
## Design primers in-house for *Botryllus schlosseri*.
### Protocol
To design primers in-house for *Botryllus schlosseri* we will utilize the [Botryllus Genome Project](http://botryllus.stanford.edu/botryllusgenome/) browser. Here we will use their BLAST tool to search for hits in query sequences as well as use their available sequecnced genome to extract those sequences which will allow us to design primers in IDT's [PrimerQuest](https://www.idtdna.com/pages/tools/primerquest?utm_source=google&utm_medium=cpc&utm_campaign=00583_1a_03&utm_content=search&gclid=CjwKCAjw-KipBhBtEiwAWjgwrEjnuHGjASk4eOYOapQcZynQKdZeAHn36oT-N5doY-ITCkQ6zzQ_KxoCm6AQAvD_BwE).
1. **Start with Gene of Interest:** Identify your gene of interest. You might want to use annotated genes for more closely related species.
Go directly to [NCBI](https://www.ncbi.nlm.nih.gov/) select the database to be nucleotide, and paste in the following but replacing "**GOI**" with your gene of interest:
> "((**GOI**\[Botryllus schlosseri\]) AND"chordates"\[porgn:\_\_txid7711\]) AND "tunicates"\[porgn:\_\_txid7712\]"
This will pull up any related gene sequences for your gene of interest for *Botryllus schlosseri* and any related tunicate species. You may use mRNA or protein sequences for your query, just be sure to select either blastn or tblastn, respectively.
::: callout-tip
Try to select sequences to blast that are shorter and more specific, ideally no more than 1,000 bp for sequences from a different organism. I noticed that sometimes if you blast a very long sequence from a different organism, you will get E-Values that look great but could be a potentially misleading conclusion. Longer sequences have more chances to match with a target sequence, so the E-value is less informative.
:::
Try to select FASTA sequences from tunicates more closely related to *Botryllus schlosseri*. See diagram below for reference.
::: {#fig-tunicate-phylogeny layout-ncol="1"}
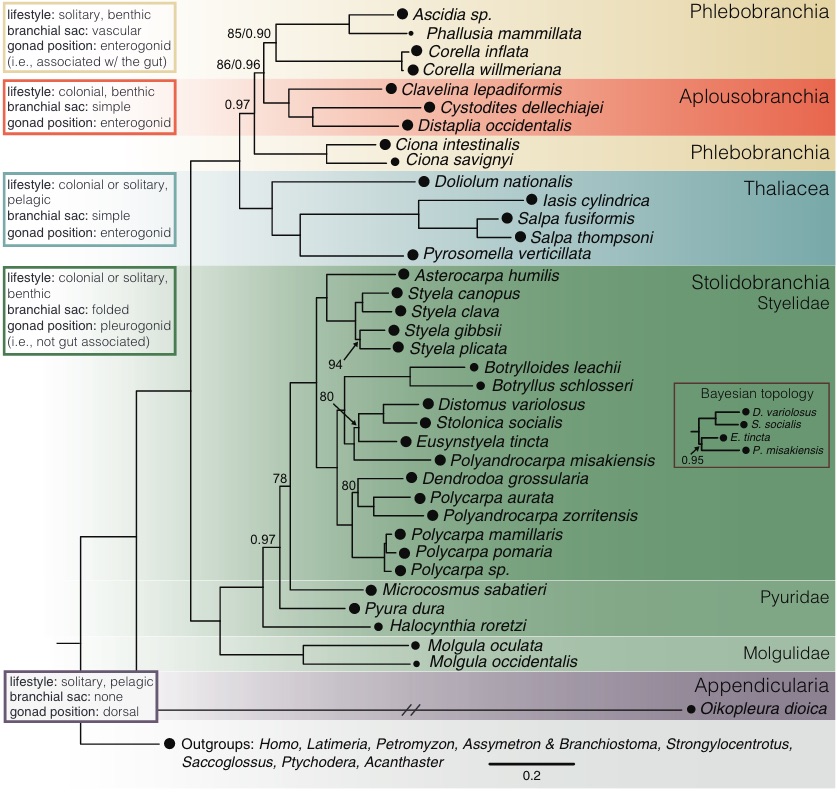
Genome Biol Evol, Volume 12, Issue 6, June 2020, Pages 948--964,
:::
2. **Retrieve Sequence:** Access the gene's sequence from NCBI Gene, by selecting the FASTA link. Copy the entire sequence starting at "\>".
3. **Use Sequence in BLAST:** Paste the sequence into the [Botryllus Genome Project's BLAST Tool](http://botryllus.stanford.edu/botryllusgenome/blast/) to identify homologous sequences in the *Botryllus* genome.
4. **Select Top Sequence:** Identify the top sequence from the BLAST results, which has a significant alignment to your gene of interest. You will only want to pursue top hits that have an E-Value of ≤ 1e-10. Record the sequence ID, E-Value, and Score (bits).
::: callout-tip
Pay attention to the E value and Score (bits) to determine if your top hit sequence for the subject (*Botryllus* contig database) is significantly in alignment with the query. A commonly used threshold is E ≤ 0.001, which suggests that you would expect to see a similar match in a random database of the same size less than once by chance. **For more stringent analyses where you want highly reliable matches, you can set a lower E-value threshold, such as E ≤ 1e-5 (0.00001) or even E ≤ 1e-10 (0.0000000001).**
:::
5. **Retrieve Botryllus Sequence:** You locate the matching sequence from the *Botryllus* genome by searching the "botznik-ctg.fa" file using the contig name you found.
6. **Primer Design:** Finally, we will use the extracted *Botryllus* sequence to design primers. We will design primers using [IDT's PrimerQuest Tool](https://www.idtdna.com/PrimerQuest/Home/Index).
::: callout-note
We will be using SYBR Green for the qPCR we are conducting so when on PrimerQuest select **qPCR Intercalating Dyes (Primers only)**
:::
This method allows us to leverage well-annotated gene data to find potential homologs in *Botryllus schlosseri*, even when the genome may not be fully annotated. We will want to validate the primers and ideally carry out experimental verification to confirm the presence and function of the genes in *Botryllus schlosseri*.
#### Sequences Identified with BLAST Tool in the botznik-ctg.fa Database
| Role | Gene ID | Query NCBI GenBank Accession | Query Organism | Top Sequence Producing Significant Alignment | Score (bits) | E Value |
|:----------|:----------|:----------|:----------|:----------|:----------|:----------|
| Apoptosis | TNF | LR791346.1 | \[Phallusia mammillata\] | botctg065850 | 70 | 1.00E-09 |
| DNA Repair | PolB | LR789137.1 | \[Phallusia mammillata\] | botctg092056 | 60 | 1.00E-06 |
| Oxidative Stress | SOD1 | XP_039262007.1 | \[Styela clava\] | botctg006757 | 199 | 8.00E-51 |
| Senescence, DNA Damage Repair | SIRT6 | BAO57219.1 | \[Polyandrocarpa misakiensis\] | botctg109566 | 446 | e-124 |
| Apoptosis | BAX | KU948200.1 | \[Botryllus schlosseri\] | botctg061218 | 2260 | 0 |
| Apoptosis | AIF1 | KU948202 | \[Botryllus schlosseri\] | botctg003666 | 674 | 0 |
| Apoptosis | IAP7 | KU948203.1 | \[Botryllus schlosseri\] | botctg106479 | 1483 | 0 |
| DNA repair | PARP1 | KU948201.1 | \[Botryllus schlosseri\] | botctg019995 | 1191 | 0 |
| Stemness | MYC | OL828248.1 | \[Botryllus schlosseri\] | botctg007779 | 2857 | 0 |
| Stemness | SOXB1 | OL828250.1 | \[Botryllus schlosseri\] | botctg003503 | 1173 | 0 |
| Stemness | POU3 | OL828252.1 | \[Botryllus schlosseri\] | botctg006949 | 1383 | 0 |
#### Pulling *Botryllus schlosseri* Sequences and Placing in New File
In order to access the sequences for the genes of interest (GOI) it will be easier for us to curl the botryllus contig sequences fasta file now available on the [Botryllus Genome Project](http://botryllus.stanford.edu/botryllusgenome/) website. I selected this file as opposed to the complete transcripts file since upon initial investigation, I found that it was more complete than the transcripts file. Because the Botryllus schlosseri transcriptome is not well-annotated, the contig sequences file may allow me to identify potentially novel genes.
```{r curl contig file, engine='bash', cache = TRUE, eval = FALSE}
curl -o data/botznik-ctg.fa http://botryllus.stanford.edu/botryllusgenome/download/botznik-ctg.fa
```
Let's take a peak at the file we just downloaded.
```{r peak at contig file, engine='bash', cache = TRUE}
head -n 10 data/botznik-ctg.fa
```
Create an empty extracted_sequences.csv file. We will upload this csv file to PrimerQuest directly.
```{r creates empty fasta and csv files, engine ='bash', eval = FALSE}
touch output/extracted_sequences.csv
```
Now let's pull the sequence for the top hit contig. Below the code will check if the sequence identifier already exists in the extracted_sequenences.csv file prior to reprinting it into the file. \]
This will make us a csv file that we can upload to PrimerQuest directly.
```{r csv file for extracted sequences, engine = 'bash', cache = TRUE, eval = FALSE}
# Define a mapping of identifiers to gene names
declare -A gene_map
gene_map[">botctg065850"]="TNF"
gene_map[">botctg092056"]="PolB"
gene_map[">botctg006757"]="SOD1"
gene_map[">botctg109566"]="SIRT6"
gene_map[">botctg061218"]="BAX"
gene_map[">botctg003666"]="AIF1"
gene_map[">botctg106479"]="IAP7"
gene_map[">botctg019995"]="PARP1"
gene_map[">botctg007779"]="MYC"
gene_map[">botctg003503"]="SOXB1"
gene_map[">botctg006949"]="POU3"
# Output CSV file
output_file="output/extracted_sequences.csv"
# Add column headers
echo "SequenceName,Sequence" > "$output_file"
# Loop through the identifiers
for identifier in "${!gene_map[@]}"; do
gene_name="${gene_map[$identifier]}"
# Extract the sequence and append it to the CSV file
awk -v id="$identifier" -v gene="$gene_name" '$0 ~ id {flag=1; id_and_gene = gene; next} /^>/{if (flag) print id_and_gene "," seq; flag=0} flag {seq = seq $0}' data/botznik-ctg.fa >> "$output_file"
done
```
Let's check out the extracted sequences in the csv file:
```{r, cache=TRUE}
read.csv(file = "output/extracted_sequences.csv")
```
#### Designing Primers with IDT PrimerQuest
General rules for designing good primers:
- 20 nucleotides in length, ideally
- 50% GC content, ideally
- Tm's of forward and reverse primers must be similar (2oC differences tolerated).
- Primers cannot form stem-loops upon themselves.
- No primer-dimers greater than 6 bp between each primer and itself and also between the two primers.
- Forward and reverse primers should be about 500 bp apart.
- The 3' end of the primer should be a G or a C.
# Confirmed Primers
| Gene | Designer | NCBI Accession Number | Fwd Primer | Rev Primer |
|:--------------|:--------------|:--------------|:--------------|:--------------|
| Mortalin | GJ | KY212120 | TTCATGATACCGAA
ACCAAGATGGACG | AATTTCCTCCTTG
ATGGCATCCACC |
| HSP70 | GJ | TBD | TBD | TBD |
# Sandbox Section
Newly released [annotated genome for botryllus schlosseri](https://aniseed.fr/aniseed/species/show_species?unique_name=Botryllus_schlosseri).
Let's curl in the genome and the functional annotations.
```{r curl contig file, engine='bash', cache = TRUE, eval = FALSE}
wget -o data/botznik-ctg.fa https://aniseed.fr/aniseed/download/?file=data%2Fboschl%2Fbotryllus_transcripts_gff_fasta.zip
```
Let's take a peak at the file we just downloaded.
```{r peak at contig file, engine='bash', cache = TRUE}
head -n 10 data/botznik-ctg.fa
```