---
author: Sam White
toc-title: Contents
toc-depth: 5
toc-location: left
layout: post
title: PCR - Crassostrea gigas and sikamea Mantle gDNA from Marinelli Shellfish Company
date: '2019-12-03 15:30'
tags:
- PCR
- gel
- Crassostrea gigas
- Crassostrea sikamea
- Pacific oyster
- Kumamoto oyster
- gDNA
categories:
- 2019
- Miscellaneous
---
I ran this PCR a couple of times before and, embarrassingly, [I had ordered/used the wrong primers](https://robertslab.github.io/sams-notebook/posts/2019/2019-11-21-PCR---Crassostrea-gigas-and-sikamea-Mantle-gDNA-from-Marinellie-Shellfish-Company---No-Multiplex/).
Well, I ordered the correct universal cytochrome oxidase primers and used those!
| SR ID | Primer Name | Sequence |
|-------|-------------|----------------------------|
| 1739 | HC02198 | taaacttcagggtgaccaaaaaatca |
| 1738 | LCO1490 | ggtcaacaaatcataaagatattgg |
| 1736 | COCsi546r | AAGTAACCTTAATAGATCAGGGAACC |
| 1735 | COCgi269r | TCGAGGAAATTGCATGTCTGCTACAA |
Primers and cycling parameters were taken from this publication:
- [Haiyan Wang and Ximing Guo "Identification of Crassostrea ariakensis and Related Oysters by Multiplex Species-Specific PCR," Journal of Shellfish Research 27(3), 481-487, (1 May 2008).](https://www.researchgate.net/profile/Ximing_Guo/publication/259643859_Identification_of_Crassostrea_ariakensis_and_related_oysters_by_multiplex_species-specific_PCR/links/55c79eb708aeb9756746e35e/Identification-of-Crassostrea-ariakensis-and-related-oysters-by-multiplex-species-specific-PCR.pdf)
Universal cytochrome oxidase primers were from this paper:
- [Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek. 1994. DNA
primers for amplification of mitochondrial cytochrome C oxidase
subunit I from diverse metazoan invertebrate. Mol. Mar. Biol.
Biotechnol. 3:294–299.](https://www.researchgate.net/publication/15316743_DNA_primers_for_amplification_of_mitochondrial_Cytochrome_C_oxidase_subunit_I_from_diverse_metazoan_invertebrates)
This is a multiplex PCR, where the HC02198 and LCO1490 primers should amplify any _Crassostrea spp._ DNA (i.e. a positive control - 697bp) and the other two primers will amplify either _C.gigas_ (Cgi269r - 269bp) or _C.sikamea_ (Csi546r - 546bp).
Master mix calcs:
| Component | Single Rxn Vol. (uL) | Num. Rxns | Total Volumes (uL) |
|------------------------|----------------------|-----------|---------------------------|
| DNA | 4 | NA | NA |
| 2x Apex Master Mix | 12.5 | 18 | 225 |
| HC02198 (100uM) | 0.15 | 18 | 2.7 |
| LCO1490 (100uM) | 0.15 | 18 | 2.7 |
| COCgi269r (100uM) | 0.1 | 18 | 1.8 |
| COCsi546r (100uM) | 0.1 | 18 | 1.8 |
| H2O | 8 | 18 | 144 |
| | 25 | | Add 21uL to each PCR tube |
Cycling params:
95oC for 10mins
30 cycles of:
- 95oC 1min
- 51oC 1min
- 72oC 1min
72oC 10mins
PCR reactions were run on a 1.5% agarose, 1x low TAE gel with ethidium bromide.
Used the GeneRuler DNA Ladder Mix (ThermoFisher) for all gels:
---
# RESULTS
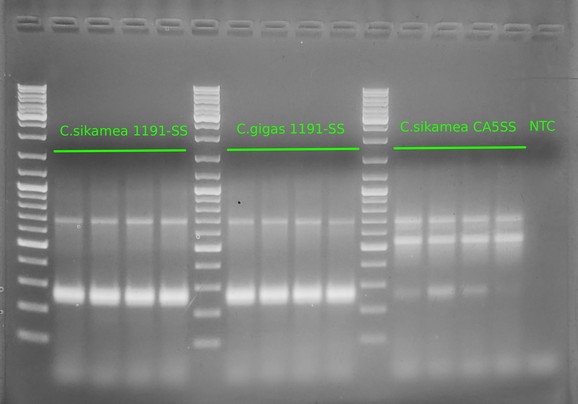
Alrighty, this is what we expected to see (at least in terms of primer functionality). We see:
- A band of ~700bp in all samples (i.e. the universal cytochrome oxidase primers)
- A band of ~270bp in the`C.gigas 1911SS` samples
Now, this is where things get interesting...
In the `C.sikamea CA5SS`:
- A prominent band of ~550bp, indicating these are _C.sikamea_
- A less prominent, but definitive, band at ~270bp, indicating the presence of _C.gigas_ cytochrome oxidase sequence. Suggests that this particular stock has hybridized with _C.gigas_ (or, there was some sort of cross contamination of DNA/tissue; unlikely as we _don't_ see potential cross contamination of _C.sikamea_ DNA in the other samples).
In the `C.sikamea 1911SS`:
- A prominent band of ~270bp, indicating these are _C.gigas_, despite the bag label indicating them as Kumamoto _and/or_ Pacific oysters (see image below)
