---
author: Sam White
toc-title: Contents
toc-depth: 5
toc-location: left
layout: post
title: PCR - Crassostrea gigas and sikamea Mantle gDNA from Marinellie Shellfish Company - No Multiplex
date: '2019-11-21 15:04'
tags:
- Crassostrea gigas
- Crassostrea sikamea
- Pacific oyster
- Kumamoto
- PCR
- Apex
- gel
categories:
- 2019
- Miscellaneous
---
### UPDATE 20191125
Since the results I obtained on my [final attempt to get this to work](https://robertslab.github.io/sams-notebook/posts/2019/2019-11-21-PCR---Crassostrea-gigas-and-sikamea-Mantle-gDNA-from-Marinellie-Shellfish-Company---No-Multiplex/) failed, I decided to double-check the primer sequences.
Well, I ordered/used the wrong sequences! The two general _Crassostrea spp._ primers ordered were the 28s primers listed in that paper, instead of the cytochrome oxidase primers! I've ordered the correct universal CO primers [which are actually listed in this paper: ([Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek. 1994. DNA
primers for amplification of mitochondrial cytochrome C oxidase
subunit I from diverse metazoan invertebrate. Mol. Mar. Biol.
Biotechnol. 3:294–299.](https://www.researchgate.net/publication/15316743_DNA_primers_for_amplification_of_mitochondrial_Cytochrome_C_oxidase_subunit_I_from_diverse_metazoan_invertebrates)) and will re-run this.
I'm leaving the original post below for posterity.
---
Primers and cycling parameters were taken from this publication:
- [Haiyan Wang and Ximing Guo "Identification of Crassostrea ariakensis and Related Oysters by Multiplex Species-Specific PCR," Journal of Shellfish Research 27(3), 481-487, (1 May 2008).](https://www.researchgate.net/profile/Ximing_Guo/publication/259643859_Identification_of_Crassostrea_ariakensis_and_related_oysters_by_multiplex_species-specific_PCR/links/55c79eb708aeb9756746e35e/Identification-of-Crassostrea-ariakensis-and-related-oysters-by-multiplex-species-specific-PCR.pdf)
| SR ID | Primer Name | Sequence |
|-------|-------------|----------------------------|
| 1727 | COreverse | CAGGGGGCCGTTCGCGGTCAACGCT |
| 1726 | COCsi546r | AAGTAACCTTAATAGATCAGGGAACC |
| 1725 | COCgi269r | TCGAGGAAATTGCATGTCTGCTACAA |
| 1724 | COforward | GGGACTACCCCCTGAATTTAAGCAT |
Instead of running a multiplex PCR as before, I ran each set of species-specific primer pairs independently.
The COforward/reverse primers should amplify any _Crassostrea spp._ DNA (i.e. a positive control - 697bp) and the other two primers will amplify either _C.gigas_ (Cgi269r - 269bp) or _C.sikamea_ (Csi546r - 546bp).
Master mix calcs:
| Component | Single Rxn Vol. (uL) | Num. Rxns | Total Volumes (uL) |
|------------------------|----------------------|-----------|---------------------------|
| DNA | 4 | NA | NA |
| 2x Apex Master Mix | 12.5 | 18 | 225 |
| COforward (100uM) | 0.15 | 18 | 2.7 |
| reverse primer (100uM) | 0.10 | 18 | 1.8 |
| H2O | 8.25 | 18 | 148.5 |
| | 25 | | Add 21uL to each PCR tube |
Cycling params:
95oC for 10mins
30 cycles of:
- 95oC 1min
- 51oC 1min
- 72oC 1min
72oC 10mins
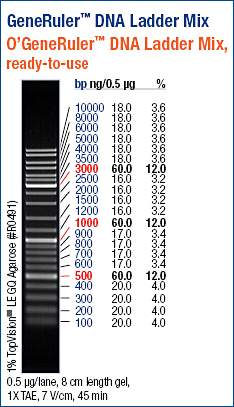
Used the GeneRuler DNA Ladder Mix (ThermoFisher) for all gels:
---
# RESULTS
GEL: _C.gigas_-specific primers:

---
GEL: _C.sikamea_-specific primers:
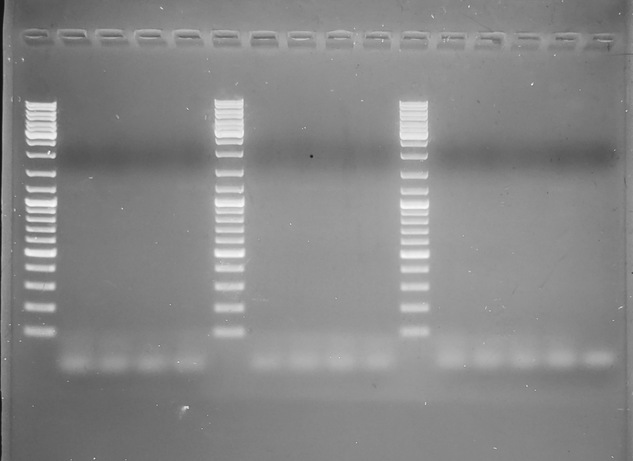
---
GEL: _Crassostrea spp._ primers:
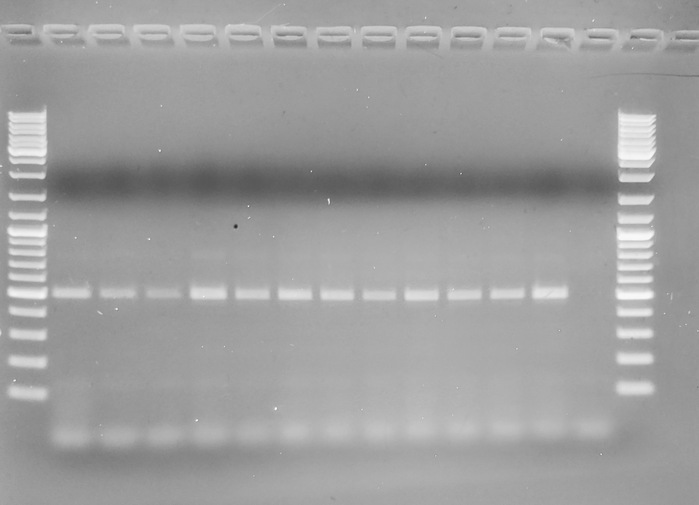
---
Well, it looks like the two species-specific reverse primers don't seem to work with the _Crassostrea spp._ forward primer.
However, the _Crassostrea spp._, despite the fact that they function properly (in that they amplify in all _Crassostrea_ DNA), they don't produce the expected band size that's mentioned in the paper (697bp). Talking with Steven, I don't think we're going to pursue much more of this.