---
author: Sam White
toc-title: Contents
toc-depth: 5
toc-location: left
layout: post
title: DNA Isolation - C.gigas Ploidy Experiment Ctenidia
date: '2019-01-17 12:45'
tags:
- ctenidia
- Crassostrea gigas
- Pacific oyster
- diploid
- triploid
- desiccation
- ploidy
categories:
- 2019
- Miscellaneous
---
Isolated DNA from the remaining ctenidia tissue samples from [Ronit's experiment](https://docs.google.com/spreadsheets/d/17mv8gMbmaldggA8Zf0RwBeNF_O4faY8dJFg31XO63K4/edit?usp=sharing) (Google Sheet).
Tissue was excised from frozen tissue block via razor blade (weight not recorded) and pulverized under liquid nitrogen. Samples were incubated O/N @ 37oC (heating block) in 350uL of MB1 Buffer + 25uL Proteinase K, per the E.Z.N.A. Mollusc DNA Kit (Omega) instructions.
After the O/N incubation, I processed the samples according to the E.Z.N.A. Mollusc DNA Kit (Omega) with the following notes:
Samples were eluted in 150uL of Elution Buffer.
Samples were stored in "[Ronit's gDNA Box #1 (positions A1 - B8 and C1 - E4)](https://docs.google.com/spreadsheets/d/1Gp91BJ5g2W6c4r8UhGVc2UhQGdCHju72aZswpR-Qxpc/edit?usp=sharing)" in the FTR213 -20oC freezer.
See either of the spreadsheets for the full list of samples isolated today.
---
NOTE: One sample (D18) did not isolate properly. After the chloroform:IAA treatment, the sample did not produce an aqueous phase (even after the specified troubleshooting step in the manual). I will have to perform another isolation on this sample next week.
Here's an image of how the sample appeared:
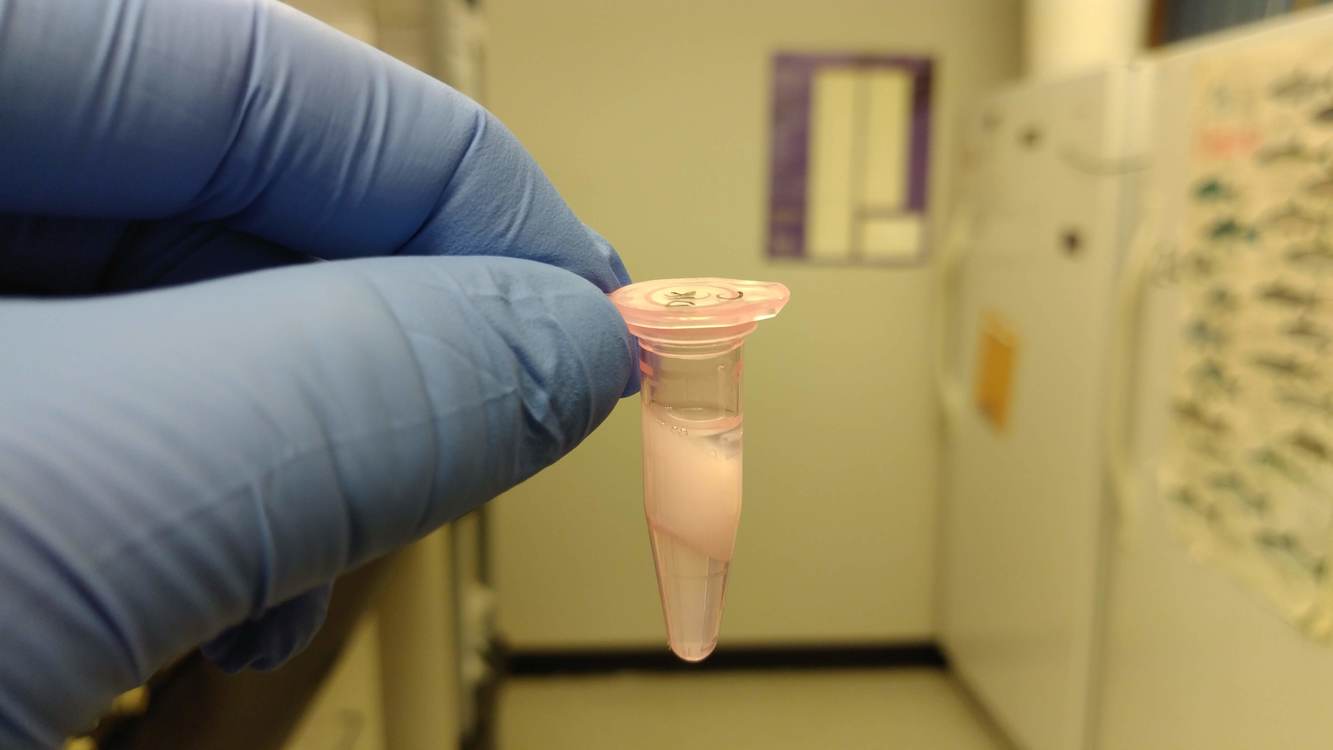
That white "fluffy" interphase should be highly compressed in a very tight, flat, smooth layer separating the organic (bottom clear liquid) and the aqueous (top clear liquid) phases.