---
title: "01-Enrichment"
output: html_document
---
Perform GO enrichment on the "top marker" genes for each partition. The background for all genes is in the "all genes" link. The marker genes (top 25, 40 and 80 genes) are also provided. For the marker genes, enrichment should be performed on each partition separately. In these files the partition is labeled "cell_group". The gene ID is listed in the column "gene_id".
all genes: https://gannet.fish.washington.edu:5001/sharing/Ra8F2cH1C
top25: https://gannet.fish.washington.edu:5001/sharing/4VIKzHDQG
top40: https://gannet.fish.washington.edu:5001/sharing/uwgGa8aPD
top80: https://gannet.fish.washington.edu:5001/sharing/L7YvQz2tk
in ../data/
```{r}
library(tidyverse)
```
```{r}
top80 <- read.csv("../data/gast_topmarkers_annot_top80.txt", sep = "\t", header = TRUE)
```
```{r}
head(top80)
```
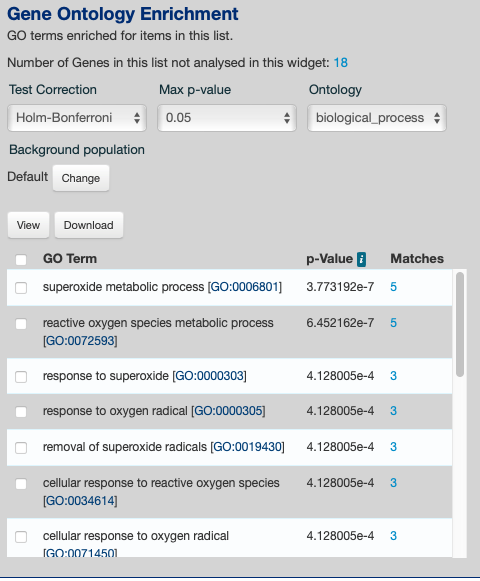
Will try cell group 1 in Aquamine
Was able to get some info with L. giantea
https://d.pr/i/GjDSk7
```{bash}
head ../output/Lgig_top80_group01.tsv
```
```{python}
from __future__ import print_function
from intermine.webservice import Service
service = Service("https://aquamine.rnet.missouri.edu/aquamine/service", token = "YOUR-API-KEY")
query = service.new_query("Gene")
query.add_view(
"primaryIdentifier", "source", "biotype", "symbol", "name", "length",
"chromosome.primaryIdentifier", "chromosomeLocation.start",
"chromosomeLocation.end", "chromosomeLocation.strand", "organism.shortName",
"chromosome.assembly"
)
query.add_constraint("Gene", "IN", "L. gigantea orthologues of TOP_80-group01_3", code="A")
for row in query.rows():
print(row["primaryIdentifier"], row["source"], row["biotype"], row["symbol"], row["name"], \
row["length"], row["chromosome.primaryIdentifier"], row["chromosomeLocation.start"], \
row["chromosomeLocation.end"], row["chromosomeLocation.strand"], row["organism.shortName"], \
row["chromosome.assembly"])
```
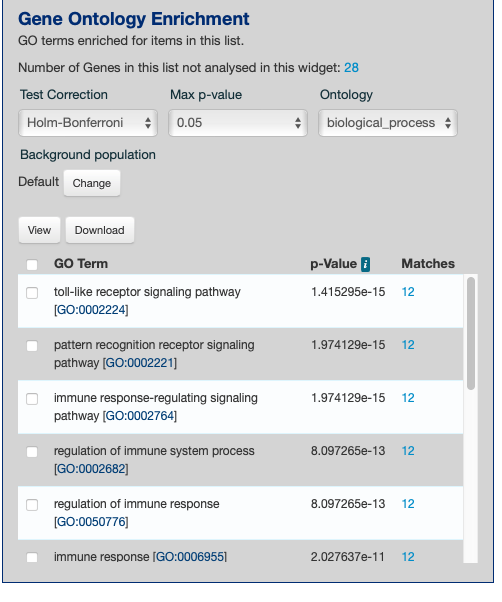
group 02
```{r}
top25 <- read.csv("../data/gast_topmarkers_annot_top25.txt", sep = "\t", header = TRUE)
```
```{r}
library(clipr)
library(xclip)
```
```{r}
head(top25)
```
```{r}
t25.2 <- dplyr::filter(top25, cell_group == "2") %>% select("gene_id")
```
```{bash}
xclip
```