DML using methylkit
================
Zoe Dellaert
10/02/2024
I am identifying differentially methylated loci using methylkit based on
[Yaamini Venkataraman’s code](https://osf.io/u46xj)
# Prepare R Markdown file
# Install packages to run methylkit
``` r
#Packages for running methylKit
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
#BiocManager::install("methylKit")
require(methylKit)
```
## Loading required package: methylKit
## Loading required package: GenomicRanges
## Loading required package: stats4
## Loading required package: BiocGenerics
##
## Attaching package: 'BiocGenerics'
## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs
## The following objects are masked from 'package:base':
##
## anyDuplicated, aperm, append, as.data.frame, basename, cbind,
## colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
## get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
## match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
## Position, rank, rbind, Reduce, rownames, sapply, setdiff, table,
## tapply, union, unique, unsplit, which.max, which.min
## Loading required package: S4Vectors
##
## Attaching package: 'S4Vectors'
## The following object is masked from 'package:utils':
##
## findMatches
## The following objects are masked from 'package:base':
##
## expand.grid, I, unname
## Loading required package: IRanges
## Loading required package: GenomeInfoDb
``` r
#install.packages("tidyverse")
require(tidyverse)
```
## Loading required package: tidyverse
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.5
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 3.5.1 ✔ tibble 3.2.1
## ✔ lubridate 1.9.3 ✔ tidyr 1.3.1
## ✔ purrr 1.0.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ lubridate::%within%() masks IRanges::%within%()
## ✖ dplyr::collapse() masks IRanges::collapse()
## ✖ dplyr::combine() masks BiocGenerics::combine()
## ✖ dplyr::desc() masks IRanges::desc()
## ✖ tidyr::expand() masks S4Vectors::expand()
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::first() masks S4Vectors::first()
## ✖ dplyr::lag() masks stats::lag()
## ✖ ggplot2::Position() masks BiocGenerics::Position(), base::Position()
## ✖ purrr::reduce() masks GenomicRanges::reduce(), IRanges::reduce()
## ✖ dplyr::rename() masks S4Vectors::rename()
## ✖ lubridate::second() masks S4Vectors::second()
## ✖ lubridate::second<-() masks S4Vectors::second<-()
## ✖ dplyr::select() masks methylKit::select()
## ✖ dplyr::slice() masks IRanges::slice()
## ✖ tidyr::unite() masks methylKit::unite()
## ℹ Use the conflicted package () to force all conflicts to become errors
``` r
#Packages for visualization
#install.packages("vegan")
#install.packages("gplots")
#install.packages("dichromat")
require(vegan)
```
## Loading required package: vegan
## Loading required package: permute
## Loading required package: lattice
## This is vegan 2.6-8
``` r
require(gplots)
```
## Loading required package: gplots
##
## Attaching package: 'gplots'
## The following object is masked from 'package:IRanges':
##
## space
## The following object is masked from 'package:S4Vectors':
##
## space
## The following object is masked from 'package:stats':
##
## lowess
``` r
require(dichromat)
```
## Loading required package: dichromat
``` r
require(readr)
```
# Obtain session information
``` r
sessionInfo()
```
## R version 4.4.0 (2024-04-24)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 22.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Etc/UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] dichromat_2.0-0.1 gplots_3.1.3.1 vegan_2.6-8
## [4] lattice_0.22-6 permute_0.9-7 lubridate_1.9.3
## [7] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
## [10] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1
## [13] tibble_3.2.1 ggplot2_3.5.1 tidyverse_2.0.0
## [16] methylKit_1.30.0 GenomicRanges_1.56.1 GenomeInfoDb_1.40.1
## [19] IRanges_2.38.1 S4Vectors_0.42.1 BiocGenerics_0.50.0
## [22] BiocManager_1.30.23
##
## loaded via a namespace (and not attached):
## [1] bitops_1.0-8 rlang_1.1.3
## [3] magrittr_2.0.3 matrixStats_1.4.1
## [5] compiler_4.4.0 mgcv_1.9-1
## [7] vctrs_0.6.5 reshape2_1.4.4
## [9] pkgconfig_2.0.3 crayon_1.5.2
## [11] fastmap_1.1.1 XVector_0.44.0
## [13] caTools_1.18.3 utf8_1.2.4
## [15] Rsamtools_2.20.0 rmarkdown_2.26
## [17] tzdb_0.4.0 UCSC.utils_1.0.0
## [19] xfun_0.43 zlibbioc_1.50.0
## [21] jsonlite_1.8.8 DelayedArray_0.30.1
## [23] BiocParallel_1.38.0 parallel_4.4.0
## [25] cluster_2.1.6 R6_2.5.1
## [27] stringi_1.8.4 limma_3.60.5
## [29] rtracklayer_1.64.0 numDeriv_2016.8-1.1
## [31] Rcpp_1.0.12 SummarizedExperiment_1.34.0
## [33] knitr_1.46 R.utils_2.12.3
## [35] Matrix_1.7-0 splines_4.4.0
## [37] timechange_0.3.0 tidyselect_1.2.1
## [39] qvalue_2.36.0 rstudioapi_0.16.0
## [41] abind_1.4-5 yaml_2.3.8
## [43] codetools_0.2-20 curl_5.2.1
## [45] plyr_1.8.9 Biobase_2.64.0
## [47] withr_3.0.0 coda_0.19-4.1
## [49] evaluate_0.23 mclust_6.1.1
## [51] Biostrings_2.72.1 pillar_1.9.0
## [53] MatrixGenerics_1.16.0 KernSmooth_2.23-22
## [55] generics_0.1.3 RCurl_1.98-1.16
## [57] emdbook_1.3.13 hms_1.1.3
## [59] munsell_0.5.1 scales_1.3.0
## [61] gtools_3.9.5 glue_1.7.0
## [63] tools_4.4.0 BiocIO_1.14.0
## [65] data.table_1.15.4 GenomicAlignments_1.40.0
## [67] mvtnorm_1.2-4 XML_3.99-0.16.1
## [69] grid_4.4.0 bbmle_1.0.25.1
## [71] bdsmatrix_1.3-7 colorspace_2.1-0
## [73] nlme_3.1-164 GenomeInfoDbData_1.2.12
## [75] restfulr_0.0.15 cli_3.6.2
## [77] fastseg_1.50.0 fansi_1.0.6
## [79] S4Arrays_1.4.1 gtable_0.3.5
## [81] R.methodsS3_1.8.2 digest_0.6.35
## [83] SparseArray_1.4.8 rjson_0.2.23
## [85] htmltools_0.5.8.1 R.oo_1.26.0
## [87] lifecycle_1.0.4 httr_1.4.7
## [89] statmod_1.5.0 MASS_7.3-60.2
# load in files from bismark
``` r
analysisFiles <- as.list(list.files("../output/05-methylKit/merged_cpg", full.names=T)) #Put all .cov files into a list for analysis.
head(analysisFiles)
```
## [[1]]
## [1] "../output/05-methylKit/merged_cpg/1.CpG_report.merged_CpG_evidence.cov.gz"
##
## [[2]]
## [1] "../output/05-methylKit/merged_cpg/10.CpG_report.merged_CpG_evidence.cov.gz"
##
## [[3]]
## [1] "../output/05-methylKit/merged_cpg/11.CpG_report.merged_CpG_evidence.cov.gz"
##
## [[4]]
## [1] "../output/05-methylKit/merged_cpg/12.CpG_report.merged_CpG_evidence.cov.gz"
##
## [[5]]
## [1] "../output/05-methylKit/merged_cpg/13.CpG_report.merged_CpG_evidence.cov.gz"
##
## [[6]]
## [1] "../output/05-methylKit/merged_cpg/14.CpG_report.merged_CpG_evidence.cov.gz"
``` r
metadata_WGBS <- read_csv("../data/metadata.WGBS.csv")
```
## Rows: 32 Columns: 4
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (2): fragment.ID, treatment
## dbl (2): block, Sample.ID
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
``` r
# Extract numbers from bam_list filenames
file_numbers <- gsub("^.*\\/([0-9]+)\\.CpG_report.*$", "\\1", analysisFiles)
file_numbers
```
## [1] "1" "10" "11" "12" "13" "14" "15" "16" "17" "18" "19" "2" "20" "21" "22"
## [16] "23" "24" "25" "26" "27" "28" "29" "3" "30" "31" "32" "4" "5" "6" "7"
## [31] "8" "9"
``` r
# Create a new dataframe with bam numbers and corresponding sample information
merged_data <- data.frame(File_Number = as.numeric(file_numbers))
# Add Sample.ID from the CSV based on matching Bam_Number to Sample.ID
merged_data$fragment.ID <- metadata_WGBS$fragment.ID[match(merged_data$File_Number, metadata_WGBS$Sample.ID)]
merged_data$treatment <- metadata_WGBS$treatment[match(merged_data$File_Number, metadata_WGBS$Sample.ID)]
# Convert treatment values: 1 for "enriched", 0 for "control"
merged_data$treatment <- ifelse(merged_data$treatment == "enriched", 1,
ifelse(merged_data$treatment == "control", 0, NA))
# Display the updated merged_data
head(merged_data)
```
## File_Number fragment.ID treatment
## 1 1 PV_3 1
## 2 10 PV_24 0
## 3 11 PV_10 1
## 4 12 PV_27 0
## 5 13 PV_22 0
## 6 14 PV_11 1
I’ll use `methRead` to create a methylation object from the coverage
files, and include sample ID and treatment information.
``` r
processedFiles <- methylKit::methRead(analysisFiles,
sample.id = as.list(file_numbers),
assembly = "Pver",
treatment = merged_data$treatment,
pipeline = "bismarkCoverage",
mincov = 2) #Process files. Use mincov = 2 to quickly process reads.
### Save the MethylKit object; re-doing the previous step is memory/time intensive, so best to use the saved object moving forward.
save(processedFiles, file = "../output/05-methylKit/processedFiles.RData")
```
``` r
load("../output/05-methylKit/processedFiles.RData")
processedFilteredFilesCov5 <- methylKit::filterByCoverage(processedFiles,
lo.count = 5, lo.perc = NULL,
high.count = NULL, high.perc = 99.9) %>%
methylKit::normalizeCoverage(.)
rm(processedFiles)
#Filter coverage information for minimum 5x coverage, and remove PCR duplicates by excluding data in the 99.9th percentile of coverage with hi.perc = 99.9. Normalize coverage between samples to avoid over-sampling reads from one sample during statistical testing
```
``` r
meth_filter=methylKit::unite(processedFilteredFilesCov5)
```
## uniting...
``` r
clusterSamples(meth_filter, dist="correlation", method="ward", plot=TRUE)
```
## The "ward" method has been renamed to "ward.D"; note new "ward.D2"
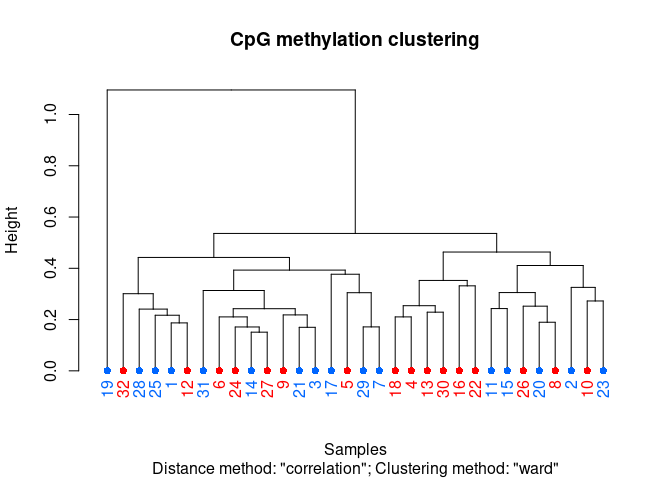
##
## Call:
## hclust(d = d, method = HCLUST.METHODS[hclust.method])
##
## Cluster method : ward.D
## Distance : pearson
## Number of objects: 32
``` r
PCASamples(meth_filter)
```
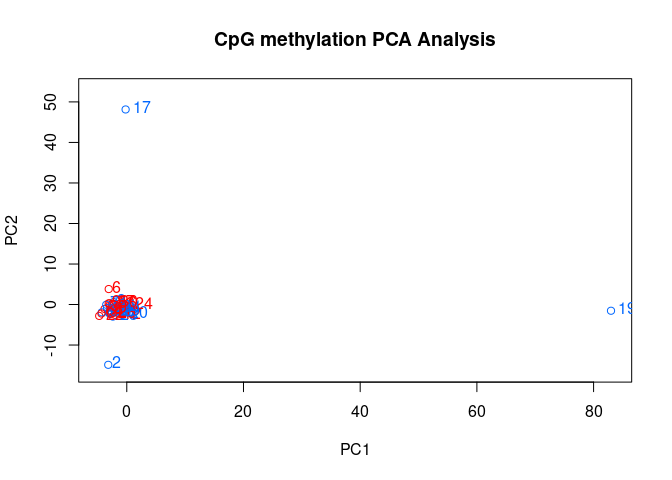
# Exclude samples 19 and 2
``` r
outliers <- c(19,2)
outliers_pos <- which(merged_data$File_Number %in% c(19,2))
outliers_frags <- merged_data$fragment.ID[which(merged_data$File_Number %in% c(19,2))]
samples_outrm <- setdiff(as.list(file_numbers),outliers)
treatment_outrm <- merged_data$treatment[-outliers_pos]
frags_outrm <- merged_data$fragment.ID[-outliers_pos]
processedFilteredFilesCov5_outrm <- methylKit::reorganize(processedFilteredFilesCov5,
sample.ids = samples_outrm,
treatment = treatment_outrm)
```
``` r
meth_filter_rmout=methylKit::unite(processedFilteredFilesCov5_outrm)
```
## uniting...
``` r
clusterSamples(meth_filter_rmout, dist="correlation", method="ward", plot=TRUE)
```
## The "ward" method has been renamed to "ward.D"; note new "ward.D2"
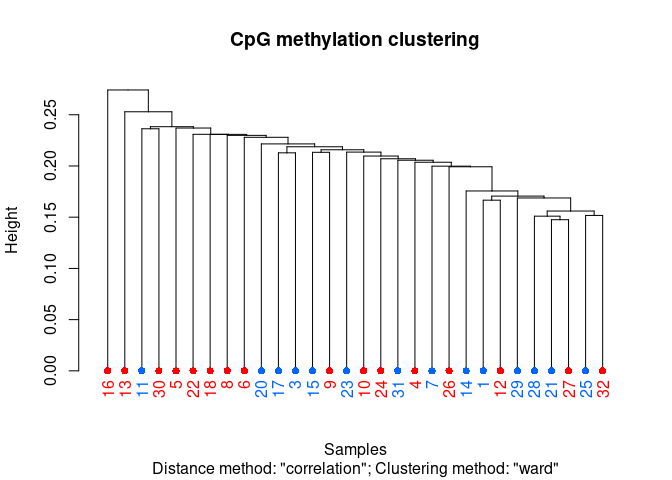
##
## Call:
## hclust(d = d, method = HCLUST.METHODS[hclust.method])
##
## Cluster method : ward.D
## Distance : pearson
## Number of objects: 30
``` r
PCASamples(meth_filter_rmout)
```
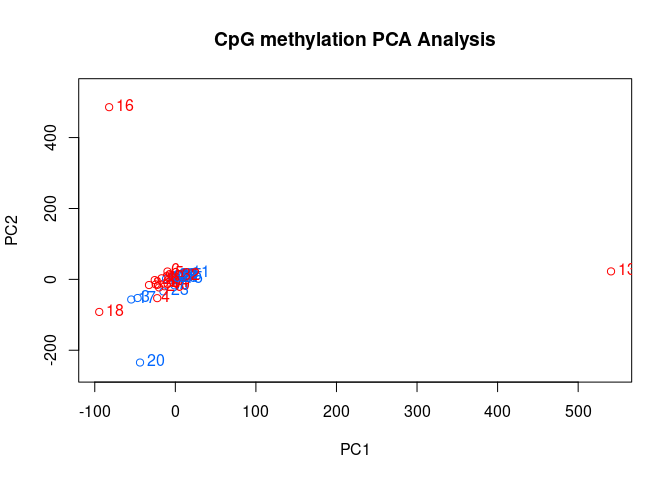
# Characterize general methylation
## Sample-specific descriptive statistics
### Specify working directory for output
``` bash
mkdir ../output/05-methylKit/general-stats
```
``` r
nFiles <- 30 #Count number of samples
fileName <- data.frame("nameBase" = rep("../output/05-methylKit/general-stats/percent-CpG-methylation", times = nFiles),
"nameBase2" = rep("../output/05-methylKit/general-stats/percent-CpG-coverage", times = nFiles),
"fragment.ID" = frags_outrm,
"File_Number" = unlist(samples_outrm),
"treatment" = treatment_outrm) #Create new dataframe for filenames.
head(fileName) #Confirm dataframe creation
```
## nameBase
## 1 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 2 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 3 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 4 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 5 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 6 ../output/05-methylKit/general-stats/percent-CpG-methylation
## nameBase2 fragment.ID
## 1 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_3
## 2 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_24
## 3 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_10
## 4 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_27
## 5 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_22
## 6 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_11
## File_Number treatment
## 1 1 1
## 2 10 0
## 3 11 1
## 4 12 0
## 5 13 0
## 6 14 1
``` r
fileName$actualFileName1 <- paste(fileName$nameBase, "-Filtered", "-5xCoverage-", fileName$fragment.ID, "_", fileName$treatment,".jpeg", sep = "") #Create a new column for the full filename for filtered + 5x coverage + specific sample's percent CpG methylation plot
fileName$actualFileName2 <- paste(fileName$nameBase2, "-Filtered", "-5xCoverage-", fileName$fragment.ID, "_",fileName$treatment,".jpeg", sep = "") #Create a new column for the full filename for filtered + 5x coverage + specific sample's percent CpG coverage plot
head(fileName) #Confirm column creation
```
## nameBase
## 1 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 2 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 3 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 4 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 5 ../output/05-methylKit/general-stats/percent-CpG-methylation
## 6 ../output/05-methylKit/general-stats/percent-CpG-methylation
## nameBase2 fragment.ID
## 1 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_3
## 2 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_24
## 3 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_10
## 4 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_27
## 5 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_22
## 6 ../output/05-methylKit/general-stats/percent-CpG-coverage PV_11
## File_Number treatment
## 1 1 1
## 2 10 0
## 3 11 1
## 4 12 0
## 5 13 0
## 6 14 1
## actualFileName1
## 1 ../output/05-methylKit/general-stats/percent-CpG-methylation-Filtered-5xCoverage-PV_3_1.jpeg
## 2 ../output/05-methylKit/general-stats/percent-CpG-methylation-Filtered-5xCoverage-PV_24_0.jpeg
## 3 ../output/05-methylKit/general-stats/percent-CpG-methylation-Filtered-5xCoverage-PV_10_1.jpeg
## 4 ../output/05-methylKit/general-stats/percent-CpG-methylation-Filtered-5xCoverage-PV_27_0.jpeg
## 5 ../output/05-methylKit/general-stats/percent-CpG-methylation-Filtered-5xCoverage-PV_22_0.jpeg
## 6 ../output/05-methylKit/general-stats/percent-CpG-methylation-Filtered-5xCoverage-PV_11_1.jpeg
## actualFileName2
## 1 ../output/05-methylKit/general-stats/percent-CpG-coverage-Filtered-5xCoverage-PV_3_1.jpeg
## 2 ../output/05-methylKit/general-stats/percent-CpG-coverage-Filtered-5xCoverage-PV_24_0.jpeg
## 3 ../output/05-methylKit/general-stats/percent-CpG-coverage-Filtered-5xCoverage-PV_10_1.jpeg
## 4 ../output/05-methylKit/general-stats/percent-CpG-coverage-Filtered-5xCoverage-PV_27_0.jpeg
## 5 ../output/05-methylKit/general-stats/percent-CpG-coverage-Filtered-5xCoverage-PV_22_0.jpeg
## 6 ../output/05-methylKit/general-stats/percent-CpG-coverage-Filtered-5xCoverage-PV_11_1.jpeg
### Create plots
``` r
for(i in 1:nFiles) { #For each data file
jpeg(filename = fileName$actualFileName1[i], height = 1000, width = 1000) #Save file with designated name
methylKit::getMethylationStats(processedFilteredFilesCov5_outrm[[i]], plot = TRUE, both.strands = FALSE) #Get %CpG methylation information
dev.off() #Turn off plotting device
} #Plot and save %CpG methylation information
```
``` r
for(i in 1:nFiles) { #For each data file
jpeg(filename = fileName$actualFileName2[i], height = 1000, width = 1000) #Save file with designated name
methylKit::getCoverageStats(processedFilteredFilesCov5_outrm[[i]], plot = TRUE, both.strands = FALSE) #Get CpG coverage information
dev.off() #Turn off plotting device
} #Plot and save CpG coverage information
```
## Comparative analysis
``` r
methylationInformationFilteredCov5 <- methylKit::unite(processedFilteredFilesCov5_outrm,
destrand = FALSE,
mc.cores = 2) #Combine all processed files into a single table. Use destrand = FALSE to not destrand. By default only bases with data in all samples will be kept
```
## uniting...
``` r
head(methylationInformationFilteredCov5) #Confirm unite
```
## chr start end strand coverage1 numCs1 numTs1
## 1 Pver_Sc0000000_size2095917 926 928 * 34 0 34
## 2 Pver_Sc0000000_size2095917 1467 1469 * 57 1 56
## 3 Pver_Sc0000000_size2095917 3870 3872 * 67 1 66
## 4 Pver_Sc0000000_size2095917 3918 3920 * 50 1 49
## 5 Pver_Sc0000000_size2095917 3921 3923 * 46 0 46
## 6 Pver_Sc0000000_size2095917 3975 3977 * 43 0 43
## coverage2 numCs2 numTs2 coverage3 numCs3 numTs3 coverage4 numCs4 numTs4
## 1 22 2 20 34 0 34 27 1 26
## 2 40 0 40 28 0 28 49 0 49
## 3 48 0 48 56 2 54 42 1 41
## 4 50 2 48 50 0 50 35 3 32
## 5 44 0 44 40 0 40 27 1 26
## 6 45 2 43 44 0 44 25 0 25
## coverage5 numCs5 numTs5 coverage6 numCs6 numTs6 coverage7 numCs7 numTs7
## 1 19 0 19 45 1 44 14 0 14
## 2 37 0 37 70 0 70 10 0 10
## 3 47 3 44 90 0 90 9 2 7
## 4 46 2 44 73 0 73 16 0 16
## 5 35 0 35 60 0 60 12 0 12
## 6 40 0 40 42 0 42 16 0 16
## coverage8 numCs8 numTs8 coverage9 numCs9 numTs9 coverage10 numCs10 numTs10
## 1 16 0 16 8 0 8 42 0 42
## 2 19 0 19 26 0 26 18 0 18
## 3 35 0 35 30 0 30 54 3 51
## 4 23 0 23 36 0 36 54 2 52
## 5 21 0 21 31 0 31 49 0 49
## 6 12 0 12 22 0 22 47 2 45
## coverage11 numCs11 numTs11 coverage12 numCs12 numTs12 coverage13 numCs13
## 1 26 0 26 27 0 27 30 0
## 2 12 0 12 47 1 46 44 0
## 3 54 0 54 47 1 46 56 2
## 4 48 2 46 40 1 39 37 0
## 5 42 0 42 35 1 34 33 0
## 6 31 0 31 28 0 28 30 0
## numTs13 coverage14 numCs14 numTs14 coverage15 numCs15 numTs15 coverage16
## 1 30 25 2 23 14 0 14 18
## 2 44 51 0 51 28 0 28 21
## 3 54 59 2 57 40 2 38 73
## 4 37 42 2 40 28 2 26 77
## 5 33 34 2 32 19 0 19 62
## 6 30 26 3 23 19 0 19 36
## numCs16 numTs16 coverage17 numCs17 numTs17 coverage18 numCs18 numTs18
## 1 0 18 30 0 30 21 0 21
## 2 0 21 48 2 46 26 1 25
## 3 1 72 79 2 77 26 0 26
## 4 1 76 64 2 62 23 0 23
## 5 1 61 40 0 40 18 0 18
## 6 0 36 40 0 40 18 1 17
## coverage19 numCs19 numTs19 coverage20 numCs20 numTs20 coverage21 numCs21
## 1 23 1 22 17 0 17 25 0
## 2 55 1 54 24 0 24 54 0
## 3 41 0 41 62 0 62 56 0
## 4 51 3 48 41 0 41 58 0
## 5 46 1 45 29 0 29 42 0
## 6 47 0 47 15 0 15 33 0
## numTs21 coverage22 numCs22 numTs22 coverage23 numCs23 numTs23 coverage24
## 1 25 16 0 16 8 0 8 18
## 2 54 9 0 9 25 0 25 30
## 3 56 21 0 21 37 0 37 35
## 4 58 14 0 14 37 0 37 36
## 5 42 14 0 14 37 0 37 32
## 6 33 18 0 18 30 0 30 24
## numCs24 numTs24 coverage25 numCs25 numTs25 coverage26 numCs26 numTs26
## 1 0 18 20 0 20 30 0 30
## 2 0 30 23 0 23 34 0 34
## 3 0 35 36 0 36 20 0 20
## 4 0 36 28 0 28 20 2 18
## 5 0 32 23 0 23 26 0 26
## 6 0 24 22 0 22 26 2 24
## coverage27 numCs27 numTs27 coverage28 numCs28 numTs28 coverage29 numCs29
## 1 32 0 32 26 0 26 9 0
## 2 38 0 38 31 0 31 9 0
## 3 22 0 22 62 0 62 16 0
## 4 34 0 34 59 0 59 23 0
## 5 32 0 32 54 0 54 33 0
## 6 24 0 24 47 2 45 24 0
## numTs29 coverage30 numCs30 numTs30
## 1 9 14 0 14
## 2 9 10 0 10
## 3 16 23 0 23
## 4 23 26 0 26
## 5 33 33 0 33
## 6 24 37 2 35
``` r
clusteringInformationFilteredCov5 <- methylKit::clusterSamples(methylationInformationFilteredCov5, dist = "correlation", method = "ward", plot = FALSE) #Save cluster information as a new object
```
## The "ward" method has been renamed to "ward.D"; note new "ward.D2"
``` r
jpeg(filename = "../output/05-methylKit/general-stats/Full-Sample-Pearson-Correlation-Plot-FilteredCov5Destrand.jpeg", height = 1000, width = 1000) #Save file with designated name
methylKit::getCorrelation(methylationInformationFilteredCov5, plot = TRUE) #Understand correlation between methylation patterns in different samples
```
## 1 10 11 12 13 14 15
## 1 1.0000000 0.8061996 0.7958106 0.8301451 0.7847248 0.8273756 0.8055034
## 10 0.8061996 1.0000000 0.7712680 0.8085445 0.7674600 0.8051597 0.7792083
## 11 0.7958106 0.7712680 1.0000000 0.7962892 0.7570843 0.7904723 0.7706376
## 12 0.8301451 0.8085445 0.7962892 1.0000000 0.7844299 0.8267122 0.8033295
## 13 0.7847248 0.7674600 0.7570843 0.7844299 1.0000000 0.7787736 0.7632122
## 14 0.8273756 0.8051597 0.7904723 0.8267122 0.7787736 1.0000000 0.7958364
## 15 0.8055034 0.7792083 0.7706376 0.8033295 0.7632122 0.7958364 1.0000000
## 16 0.7767535 0.7548201 0.7452744 0.7794010 0.7373980 0.7729037 0.7547756
## 17 0.8007072 0.7823748 0.7708605 0.8036234 0.7619920 0.7957465 0.7776216
## 18 0.7946319 0.7715894 0.7596287 0.7918804 0.7516107 0.7915973 0.7725624
## 20 0.8011124 0.7753799 0.7678618 0.8012166 0.7577290 0.7938067 0.7773504
## 21 0.8376765 0.8130425 0.8001107 0.8361573 0.7942560 0.8311890 0.8130747
## 22 0.7959472 0.7713303 0.7632831 0.7983690 0.7546978 0.7897381 0.7747832
## 23 0.8046269 0.7800467 0.7743049 0.8017605 0.7620017 0.8018848 0.7759185
## 24 0.8058729 0.7855537 0.7715400 0.8095638 0.7670965 0.8041392 0.7855213
## 25 0.8354559 0.8097619 0.7998861 0.8336079 0.7943424 0.8306403 0.8073999
## 26 0.8053940 0.7883449 0.7732101 0.8111625 0.7679860 0.8041125 0.7858633
## 27 0.8317356 0.8076019 0.7945806 0.8347240 0.7938199 0.8309117 0.8053064
## 28 0.8335141 0.8106012 0.7992608 0.8351718 0.7932822 0.8299563 0.8120815
## 29 0.8267916 0.8052480 0.7893615 0.8248645 0.7877253 0.8189214 0.8037017
## 3 0.7972643 0.7825229 0.7606560 0.7995129 0.7617854 0.7985275 0.7747318
## 30 0.7929647 0.7750965 0.7625491 0.7996153 0.7526396 0.7924026 0.7697992
## 31 0.8075267 0.7838447 0.7736599 0.8076834 0.7638965 0.8004530 0.7856930
## 32 0.8382786 0.8130856 0.8033816 0.8339395 0.7912997 0.8317501 0.8140329
## 4 0.8067752 0.7829185 0.7709134 0.8068069 0.7649580 0.8057007 0.7834105
## 5 0.7962566 0.7714665 0.7624304 0.7955222 0.7553404 0.7904131 0.7708494
## 6 0.8015575 0.7790692 0.7681564 0.8012086 0.7595051 0.7986252 0.7728562
## 7 0.8078729 0.7892221 0.7719694 0.8091966 0.7709562 0.8042977 0.7831679
## 8 0.7983097 0.7723883 0.7650225 0.7967550 0.7581625 0.7897293 0.7709289
## 9 0.8054015 0.7800769 0.7709362 0.8047559 0.7634385 0.7984601 0.7825446
## 16 17 18 20 21 22 23
## 1 0.7767535 0.8007072 0.7946319 0.8011124 0.8376765 0.7959472 0.8046269
## 10 0.7548201 0.7823748 0.7715894 0.7753799 0.8130425 0.7713303 0.7800467
## 11 0.7452744 0.7708605 0.7596287 0.7678618 0.8001107 0.7632831 0.7743049
## 12 0.7794010 0.8036234 0.7918804 0.8012166 0.8361573 0.7983690 0.8017605
## 13 0.7373980 0.7619920 0.7516107 0.7577290 0.7942560 0.7546978 0.7620017
## 14 0.7729037 0.7957465 0.7915973 0.7938067 0.8311890 0.7897381 0.8018848
## 15 0.7547756 0.7776216 0.7725624 0.7773504 0.8130747 0.7747832 0.7759185
## 16 1.0000000 0.7569781 0.7460510 0.7473816 0.7810494 0.7490368 0.7549426
## 17 0.7569781 1.0000000 0.7717849 0.7748534 0.8124896 0.7717782 0.7757378
## 18 0.7460510 0.7717849 1.0000000 0.7680225 0.8082310 0.7667937 0.7695917
## 20 0.7473816 0.7748534 0.7680225 1.0000000 0.8077659 0.7688717 0.7758778
## 21 0.7810494 0.8124896 0.8082310 0.8077659 1.0000000 0.8064314 0.8123349
## 22 0.7490368 0.7717782 0.7667937 0.7688717 0.8064314 1.0000000 0.7709144
## 23 0.7549426 0.7757378 0.7695917 0.7758778 0.8123349 0.7709144 1.0000000
## 24 0.7570280 0.7813343 0.7768157 0.7780535 0.8154307 0.7775811 0.7821289
## 25 0.7834087 0.8046682 0.8019362 0.8051814 0.8434750 0.8037176 0.8085519
## 26 0.7585795 0.7871547 0.7819335 0.7821566 0.8225009 0.7817821 0.7829239
## 27 0.7854563 0.8096996 0.8045548 0.8050802 0.8477752 0.8054932 0.8062218
## 28 0.7818233 0.8079977 0.8050945 0.8081897 0.8464504 0.8024744 0.8108802
## 29 0.7739411 0.8014435 0.8037162 0.7971029 0.8394018 0.7964799 0.7967916
## 3 0.7553582 0.7821951 0.7739470 0.7693152 0.8128327 0.7718116 0.7727913
## 30 0.7457752 0.7681297 0.7603041 0.7661190 0.8038102 0.7656733 0.7707995
## 31 0.7567794 0.7850513 0.7794980 0.7789786 0.8186137 0.7749618 0.7830781
## 32 0.7845233 0.8126823 0.8009811 0.8094919 0.8474732 0.7992072 0.8129212
## 4 0.7592794 0.7824233 0.7788728 0.7800432 0.8151053 0.7721775 0.7803325
## 5 0.7465908 0.7708721 0.7625577 0.7672779 0.8022814 0.7630086 0.7718595
## 6 0.7516654 0.7726560 0.7677232 0.7721550 0.8048218 0.7668910 0.7776883
## 7 0.7554785 0.7857897 0.7791176 0.7802455 0.8162389 0.7786752 0.7843043
## 8 0.7466012 0.7727125 0.7628209 0.7704749 0.8044546 0.7653577 0.7731016
## 9 0.7553553 0.7807637 0.7729736 0.7732094 0.8138624 0.7738779 0.7780321
## 24 25 26 27 28 29 3
## 1 0.8058729 0.8354559 0.8053940 0.8317356 0.8335141 0.8267916 0.7972643
## 10 0.7855537 0.8097619 0.7883449 0.8076019 0.8106012 0.8052480 0.7825229
## 11 0.7715400 0.7998861 0.7732101 0.7945806 0.7992608 0.7893615 0.7606560
## 12 0.8095638 0.8336079 0.8111625 0.8347240 0.8351718 0.8248645 0.7995129
## 13 0.7670965 0.7943424 0.7679860 0.7938199 0.7932822 0.7877253 0.7617854
## 14 0.8041392 0.8306403 0.8041125 0.8309117 0.8299563 0.8189214 0.7985275
## 15 0.7855213 0.8073999 0.7858633 0.8053064 0.8120815 0.8037017 0.7747318
## 16 0.7570280 0.7834087 0.7585795 0.7854563 0.7818233 0.7739411 0.7553582
## 17 0.7813343 0.8046682 0.7871547 0.8096996 0.8079977 0.8014435 0.7821951
## 18 0.7768157 0.8019362 0.7819335 0.8045548 0.8050945 0.8037162 0.7739470
## 20 0.7780535 0.8051814 0.7821566 0.8050802 0.8081897 0.7971029 0.7693152
## 21 0.8154307 0.8434750 0.8225009 0.8477752 0.8464504 0.8394018 0.8128327
## 22 0.7775811 0.8037176 0.7817821 0.8054932 0.8024744 0.7964799 0.7718116
## 23 0.7821289 0.8085519 0.7829239 0.8062218 0.8108802 0.7967916 0.7727913
## 24 1.0000000 0.8129586 0.7897985 0.8174674 0.8132591 0.8048537 0.7865854
## 25 0.8129586 1.0000000 0.8169082 0.8412878 0.8434179 0.8316815 0.8101781
## 26 0.7897985 0.8169082 1.0000000 0.8174119 0.8190821 0.8087437 0.7849661
## 27 0.8174674 0.8412878 0.8174119 1.0000000 0.8434902 0.8325710 0.8137549
## 28 0.8132591 0.8434179 0.8190821 0.8434902 1.0000000 0.8394696 0.8103390
## 29 0.8048537 0.8316815 0.8087437 0.8325710 0.8394696 1.0000000 0.8074994
## 3 0.7865854 0.8101781 0.7849661 0.8137549 0.8103390 0.8074994 1.0000000
## 30 0.7756041 0.7993107 0.7738783 0.7994793 0.8013330 0.7899446 0.7697198
## 31 0.7844874 0.8106327 0.7903634 0.8154842 0.8193549 0.8115310 0.7823082
## 32 0.8107811 0.8451294 0.8144715 0.8407032 0.8442995 0.8316885 0.8050838
## 4 0.7856437 0.8133203 0.7899371 0.8178246 0.8166103 0.8051842 0.7820797
## 5 0.7705246 0.7984390 0.7754399 0.7988042 0.7994056 0.7929591 0.7650622
## 6 0.7776868 0.8043184 0.7781655 0.8040879 0.8051958 0.7955089 0.7700254
## 7 0.7893001 0.8128983 0.7926234 0.8150149 0.8185198 0.8116655 0.7823362
## 8 0.7788059 0.8011386 0.7797101 0.8011636 0.8034457 0.7964820 0.7712334
## 9 0.7844185 0.8111096 0.7873481 0.8119523 0.8113469 0.7988477 0.7780542
## 30 31 32 4 5 6 7
## 1 0.7929647 0.8075267 0.8382786 0.8067752 0.7962566 0.8015575 0.8078729
## 10 0.7750965 0.7838447 0.8130856 0.7829185 0.7714665 0.7790692 0.7892221
## 11 0.7625491 0.7736599 0.8033816 0.7709134 0.7624304 0.7681564 0.7719694
## 12 0.7996153 0.8076834 0.8339395 0.8068069 0.7955222 0.8012086 0.8091966
## 13 0.7526396 0.7638965 0.7912997 0.7649580 0.7553404 0.7595051 0.7709562
## 14 0.7924026 0.8004530 0.8317501 0.8057007 0.7904131 0.7986252 0.8042977
## 15 0.7697992 0.7856930 0.8140329 0.7834105 0.7708494 0.7728562 0.7831679
## 16 0.7457752 0.7567794 0.7845233 0.7592794 0.7465908 0.7516654 0.7554785
## 17 0.7681297 0.7850513 0.8126823 0.7824233 0.7708721 0.7726560 0.7857897
## 18 0.7603041 0.7794980 0.8009811 0.7788728 0.7625577 0.7677232 0.7791176
## 20 0.7661190 0.7789786 0.8094919 0.7800432 0.7672779 0.7721550 0.7802455
## 21 0.8038102 0.8186137 0.8474732 0.8151053 0.8022814 0.8048218 0.8162389
## 22 0.7656733 0.7749618 0.7992072 0.7721775 0.7630086 0.7668910 0.7786752
## 23 0.7707995 0.7830781 0.8129212 0.7803325 0.7718595 0.7776883 0.7843043
## 24 0.7756041 0.7844874 0.8107811 0.7856437 0.7705246 0.7776868 0.7893001
## 25 0.7993107 0.8106327 0.8451294 0.8133203 0.7984390 0.8043184 0.8128983
## 26 0.7738783 0.7903634 0.8144715 0.7899371 0.7754399 0.7781655 0.7926234
## 27 0.7994793 0.8154842 0.8407032 0.8178246 0.7988042 0.8040879 0.8150149
## 28 0.8013330 0.8193549 0.8442995 0.8166103 0.7994056 0.8051958 0.8185198
## 29 0.7899446 0.8115310 0.8316885 0.8051842 0.7929591 0.7955089 0.8116655
## 3 0.7697198 0.7823082 0.8050838 0.7820797 0.7650622 0.7700254 0.7823362
## 30 1.0000000 0.7742005 0.8020964 0.7724805 0.7617103 0.7654493 0.7753827
## 31 0.7742005 1.0000000 0.8157225 0.7884501 0.7751963 0.7753393 0.7873818
## 32 0.8020964 0.8157225 1.0000000 0.8162836 0.8057991 0.8091238 0.8154896
## 4 0.7724805 0.7884501 0.8162836 1.0000000 0.7710843 0.7779276 0.7864458
## 5 0.7617103 0.7751963 0.8057991 0.7710843 1.0000000 0.7675738 0.7748748
## 6 0.7654493 0.7753393 0.8091238 0.7779276 0.7675738 1.0000000 0.7783753
## 7 0.7753827 0.7873818 0.8154896 0.7864458 0.7748748 0.7783753 1.0000000
## 8 0.7656478 0.7771840 0.8050747 0.7776998 0.7658136 0.7703481 0.7813665
## 9 0.7692971 0.7859809 0.8099007 0.7800522 0.7711045 0.7724759 0.7878302
## 8 9
## 1 0.7983097 0.8054015
## 10 0.7723883 0.7800769
## 11 0.7650225 0.7709362
## 12 0.7967550 0.8047559
## 13 0.7581625 0.7634385
## 14 0.7897293 0.7984601
## 15 0.7709289 0.7825446
## 16 0.7466012 0.7553553
## 17 0.7727125 0.7807637
## 18 0.7628209 0.7729736
## 20 0.7704749 0.7732094
## 21 0.8044546 0.8138624
## 22 0.7653577 0.7738779
## 23 0.7731016 0.7780321
## 24 0.7788059 0.7844185
## 25 0.8011386 0.8111096
## 26 0.7797101 0.7873481
## 27 0.8011636 0.8119523
## 28 0.8034457 0.8113469
## 29 0.7964820 0.7988477
## 3 0.7712334 0.7780542
## 30 0.7656478 0.7692971
## 31 0.7771840 0.7859809
## 32 0.8050747 0.8099007
## 4 0.7776998 0.7800522
## 5 0.7658136 0.7711045
## 6 0.7703481 0.7724759
## 7 0.7813665 0.7878302
## 8 1.0000000 0.7739608
## 9 0.7739608 1.0000000
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
## Warning in par(usr): argument 1 does not name a graphical parameter
``` r
dev.off()
```
## png
## 2
``` r
jpeg(filename = "../output/05-methylKit/general-stats/Full-Sample-CpG-Methylation-Clustering-FilteredCov5Destrand.jpeg", height = 1000, width = 1000) #Save file with designated name
methylKit::clusterSamples(methylationInformationFilteredCov5, dist = "correlation", method = "ward", plot = TRUE) #Cluster samples based on correlation coefficients
```
## The "ward" method has been renamed to "ward.D"; note new "ward.D2"
##
## Call:
## hclust(d = d, method = HCLUST.METHODS[hclust.method])
##
## Cluster method : ward.D
## Distance : pearson
## Number of objects: 30
``` r
dev.off()
```
## png
## 2
``` r
jpeg(filename = "../output/05-methylKit/general-stats/Full-Sample-Methylation-PCA-FilteredCov5Destrand.jpeg", height = 1000, width = 1000) #Save file with designated name
methylKit::PCASamples(methylationInformationFilteredCov5) #Run a PCA analysis on percent methylation for all samples
dev.off() #Turn off plotting device
```
## png
## 2
``` r
jpeg(filename = "../output/05-methylKit/general-stats/Full-Sample-Methylation-Screeplot-FilteredCov5Destrand.jpeg", height = 1000, width = 1000) #Save file with designated name
methylKit::PCASamples(methylationInformationFilteredCov5, screeplot = TRUE) #Run the PCA analysis and plot variances against PC number in a screeplot
dev.off()
```
## png
## 2
# Differentially methylated loci
## All samples
### Identify DML
``` r
differentialMethylationStatsTreatment_outrm <- methylKit::calculateDiffMeth(methylationInformationFilteredCov5, overdispersion = "MN", test = "Chisq", mc.cores = 8) #Calculate differential methylation statistics and include covariate information.
```
## two groups detected:
## will calculate methylation difference as the difference of
## treatment (group: 1) - control (group: 0)
``` r
head(differentialMethylationStatsTreatment_outrm) #Look at differential methylation output
```
## chr start end strand pvalue qvalue meth.diff
## 1 Pver_Sc0000000_size2095917 926 928 * 0.6712749 0.8780778 0.3787879
## 2 Pver_Sc0000000_size2095917 1467 1469 * 0.9014604 0.9083778 -0.0622665
## 3 Pver_Sc0000000_size2095917 3870 3872 * 0.3465944 0.7080206 -0.9553611
## 4 Pver_Sc0000000_size2095917 3918 3920 * 0.1648768 0.5428307 -1.2977170
## 5 Pver_Sc0000000_size2095917 3921 3923 * 0.1643548 0.5418196 0.6684247
## 6 Pver_Sc0000000_size2095917 3975 3977 * 0.4058801 0.7567318 -0.9086089
``` r
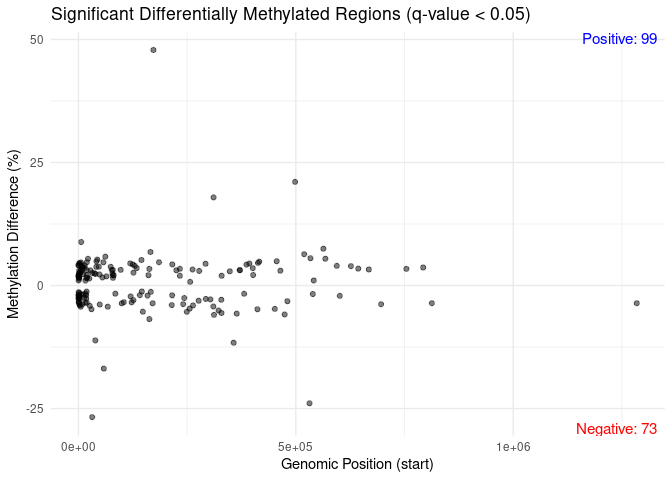
library(ggplot2)
# Assuming differentialMethylationStatsTreatment_outrm is your methylDiff object
# Filter DMRs with q-value < 0.05
significant_dmg <- getData(differentialMethylationStatsTreatment_outrm[differentialMethylationStatsTreatment_outrm$qvalue < 0.05, ])
# Create a data frame for plotting
plot_data <- data.frame(
chr = significant_dmg$chr,
start = significant_dmg$start,
meth.diff = significant_dmg$meth.diff
)
# Count the number of positive and negative methylation differences
positive_count <- sum(significant_dmg$meth.diff > 0)
negative_count <- sum(significant_dmg$meth.diff < 0)
# Plot with counts added to the quadrants
ggplot(plot_data, aes(x = start, y = meth.diff)) +
geom_point(alpha = 0.5) + # Set alpha to reduce point transparency
theme_minimal() +
labs(title = "Significant Differentially Methylated Regions (q-value < 0.05)",
x = "Genomic Position (start)",
y = "Methylation Difference (%)") +
theme(legend.position = "none") + # Remove the legend
# Add the count of positive and negative methylation differences as text annotations
annotate("text", x = Inf, y = Inf, label = paste("Positive:", positive_count),
hjust = 1.1, vjust = 1.1, size = 4, color = "blue") +
annotate("text", x = Inf, y = -Inf, label = paste("Negative:", negative_count),
hjust = 1.1, vjust = -0.1, size = 4, color = "red")
```
``` r
diffMethStatsTreatment5 <- methylKit::getMethylDiff(differentialMethylationStatsTreatment_outrm, difference = 5, qvalue = 0.01) #Identify DML based on difference threshold
```
## Warning in max(i): no non-missing arguments to max; returning -Inf
``` r
length(diffMethStatsTreatment5$chr) #DML
```
## [1] 0
``` r
head(diffMethStatsTreatment5)
```
## Warning in max(i): no non-missing arguments to max; returning -Inf
## [1] chr start end strand pvalue qvalue meth.diff
## <0 rows> (or 0-length row.names)
``` r
write.csv(diffMethStatsTreatment25All, "DML/DML-pH-25-Cov5-All.csv")
write.csv(diffMethStatsTreatment50All, "DML/DML-pH-50-Cov5-All.csv")
write.csv(diffMethStatsTreatment75All, "DML/DML-pH-75-Cov5-All.csv")
```
## Appendix
Moving Danielle files to unity from Andromeda:
in Andromeda:
``` bash
interactive
module load GlobusConnectPersonal/3.2.0
#follow the login link, enter auth code, enter "~/data/putnamlab/" as endpoint
nano ~/.globusonline/lta/config-paths
```
/data/putnamlab/,0,1 # Read-write access to the putnamlab directory
``` bash
globusconnectpersonal -start &
```
On Globus Connect in your browser (): - Login -
Select your university and login - Go to the File Manager and in the
Collection field on the left, enter the personal endpoint string that
was spit out by the globus setup above - Select
acda5457-9c06-4564-8375-260ba428f22a (exact address of Unity) in the
collection field on the right - Select the files or folders you want to
transfer from Andromeda to Unity and press ‘Start’. - navigating to
`/data/putnamlab/dbecks/Becker_E5/WGBS_Becker_E5/Becker_WGBS/bismark_deduplicated/`
in andromeda and `/project/pi_hputnam_uri_edu/WGBS_mech_model/` in
unity - navigating to
`/data/putnamlab/dbecks/Becker_E5/WGBS_Becker_E5/Becker_WGBS/CovtoCyto/`
in andromeda and
`/project/pi_hputnam_uri_edu/mechanism_to_model/output/05-methylKit/merged_cpg/`
in unity