Where are the SNPs?
================
Steven Roberts
17 July, 2023
- 1 Background
- 2 Epidiverse
- 2.1 Merging Epidiverse VCFs
- 2.1.1 Trying to
get Genetic Relatedness Matrix
- 3 BS-SNPer
- 3.1 draft paper
- 3.2 Wiki
- 3.3
issue (backtracking)
# 1 Background
At minimum, identifying C-\>T SNPs on both strands so we have a list of
SNPs from the EpiDiverse output.
Other tasks that build upon that:
- Compare Bs-snper and EpiDiverse SNP lists. Determine which source our
final C-\>T SNP list should come from
- Remove SNPs from methylKit analysis
- Remove SNPs and recalculate average gene methylation and CV of gene
methylation for modeling analyses
------------------------------------------------------------------------
There are two ways we have used to get genetic information from DNA
methylation data. - EPIDiverse - BS-SNPer
# 2 Epidiverse
Sam ran Epidiverse
- [Notebook](https://robertslab.github.io/sams-notebook/2022/09/21/BSseq-SNP-Analysis-Nextflow-EpiDiverse-SNP-Pipeline-for-C.virginica-CEABIGR-BSseq-data.html)
- VCF Directory -
- results dir -
## 2.1 Merging Epidiverse VCFs
Next step for capturing SNP info in Epidiverse Workflow is merging.
``` bash
cd ../data/big
wget -r \
--no-directories --no-parent \
-P . \
-A "*vcf.g*" https://gannet.fish.washington.edu/Atumefaciens/20220921-cvir-ceabigr-nextflow-epidiverse-snp/snps/vcf/
```
``` bash
/home/shared/bcftools-1.14/bcftools merge \
--force-samples \
../data/big/*.vcf.gz \
--merge all \
--threads 40 \
-O v \
-o ../output/51-SNPs/EpiDiv_merged.vcf
```
``` bash
head ../output/51-SNPs/EpiDiv_merged.vcf
tail -2 ../output/51-SNPs/EpiDiv_merged.vcf
```
## ##fileformat=VCFv4.2
## ##FILTER=
## ##fileDate=20220924
## ##source=freeBayes v1.3.2-dirty
## ##reference=GCF_002022765.2_C_virginica-3.0_genomic.fa
## ##contig=
## ##contig=
## ##contig=
## ##contig=
## ##contig=
## NC_007175.2 17243 . G T 24.3275 . DPB=2;EPPR=0;GTI=0;MQMR=0;NS=1;NUMALT=1;ODDS=4.55889;PAIREDR=0;PQR=0;PRO=0;QR=0;RO=0;RPPR=0;SRF=0;SRP=0;SRR=0;DP=4;AB=0;ABP=0;AF=1;AO=2;CIGAR=1X;DPRA=0;EPP=3.0103;LEN=1;MEANALT=1;MQM=21.5;PAIRED=1;PAO=0;PQA=0;QA=72;RPL=2;RPP=7.35324;RPR=0;RUN=1;SAF=1;SAP=3.0103;SAR=1;TYPE=snp;AN=4;AC=4 GT:GQ:DP:AD:RO:QR:AO:QA:GL ./.:.:.:.:.:.:.:.:. 1/1:18:2:0,2:0:0:2:48:-3.78608,-0.60206,0 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. 1/1:20:2:0,2:0:0:2:72:-3.63228,-0.60206,0 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:.
## NC_007175.2 17244 . C A,T 99.609 . DPB=18;EPPR=16.5402;GTI=0;MQMR=32.6923;NS=1;NUMALT=2;ODDS=10.1722;PAIREDR=1;PQR=0;PRO=0;QR=443;RO=13;RPPR=31.2394;SRF=11;SRP=16.5402;SRR=2;DP=636;AB=0.264706,0.22;ABP=19.3602,37.059;AF=0.5,0.5;AO=9,11;CIGAR=1X,1X;DPRA=0,0;EPP=5.18177,12.6832;LEN=1,1;MEANALT=3,2;MQM=22.5556,27.5455;PAIRED=1,1;PAO=0,0;PQA=0,0;QA=218,338;RPL=9,11;RPP=22.5536,26.8965;RPR=0,0;RUN=1,1;SAF=6,9;SAP=5.18177,12.6832;SAR=3,2;TYPE=snp,snp;AN=32;AC=9,7 GT:GQ:DP:AD:RO:QR:AO:QA:GL ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. 0/1:0:18:13,3,2:13:443:3,2:65,62:-0.3642,0,-28.1339,-0.68853,-25.4702,-29.1527 0/1:14:58:34,16,.:34:1198:16,.:373,.:-11.3765,0,-73.9751,.,.,. 0/2:27:33:15,.,10:15:543:.,10:.,289:-10.3803,.,.,0,.,-28.2326 ./.:.:.:.:.:.:.:.:. 0/1:0:14:8,3,3:8:296:3,3:56,86:-0.173115,0,-15.8277,-0.939605,-15.1184,-16.7429 ./.:.:.:.:.:.:.:.:. 0/2:94:61:31,.,19:31:1147:.,19:.,580:-19.9187,.,.,0,.,-66.5717 0/1:68:46:22,16,.:22:802:16,.:413,.:-16.0383,0,-45.3192,.,.,. 0/1:95:78:38,27,.:38:1282:27,.:642,.:-22.4786,0,-68.4162,.,.,. 0/1:64:45:18,18,.:18:654:18,.:461,.:-18.0316,0,-33.1576,.,.,. 0/1:0:20:12,5,3:12:406:5,3:117,91:-3.86175,0,-23.2009,-1.54201,-18.7955,-25.3809 0/2:0:35:23,4,7:23:801:4,7:74,209:-4.79192,-7.31508,-52.6892,0,-42.2138,-44.5142 ./.:.:.:.:.:.:.:.:. 0/2:3:27:14,.,5:14:518:.,5:.,148:-6.30832,.,.,0,.,-28.3683 0/1:0:11:7,3,.:7:259:3,.:72,.:-2.47552,0,-13.1431,.,.,. 0/2:4:52:26,.,10:26:926:.,10:.,294:-13.4825,.,.,0,.,-47.589 0/1:0:34:21,9,3:21:725:9,3:218,93:-4.45192,0,-37.6367,-3.99214,-33.5854,-43.5048 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. 0/2:92:54:29,.,14:29:1035:.,14:.,422:-18.2443,.,.,0,.,-56.7053 0/2:29:50:25,.,11:25:865:.,11:.,338:-12.416,.,.,0,.,-52.8617 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:.
``` bash
/home/shared/vcftools-0.1.16/bin/vcftools \
--vcf ../output/51-SNPs/EpiDiv_merged.vcf \
--recode --recode-INFO-all \
--min-alleles 2 --max-alleles 2 \
--max-missing 0.5 \
--mac 2 \
--out ../output/51-SNPs/EpiDiv_merged.f
```
After filtering, kept 26 out of 26 Individuals Outputting VCF file…
After filtering, kept 2343637 out of a possible 144873997 Sites Run Time
= 897.00 seconds
``` bash
head ../output/51-SNPs/EpiDiv_merged.f.recode.vcf
tail -2 ../output/51-SNPs/EpiDiv_merged.f.recode.vcf
```
## ##fileformat=VCFv4.2
## ##FILTER=
## ##fileDate=20220924
## ##source=freeBayes v1.3.2-dirty
## ##reference=GCF_002022765.2_C_virginica-3.0_genomic.fa
## ##contig=
## ##contig=
## ##contig=
## ##contig=
## ##contig=
## NC_007175.2 6500 . T A 0.198458 . DPB=199;EPPR=10.6176;GTI=0;MQMR=38.3377;NS=1;NUMALT=1;ODDS=22.6887;PAIREDR=1;PQR=0;PRO=0;QR=4953;RO=151;RPPR=79.6446;SRF=63;SRP=11.9982;SRR=88;DP=8738;AB=0;ABP=0;AF=0;AO=61;CIGAR=1X;DPRA=0;EPP=75.0961;LEN=1;MEANALT=3;MQM=27.6557;PAIRED=1;PAO=0;PQA=0;QA=1307;RPL=41;RPP=18.709;RPR=20;RUN=1;SAF=37;SAP=9.02635;SAR=24;TYPE=snp;AN=42;AC=4 GT:GQ:DP:AD:RO:QR:AO:QA:GL ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. 0/1:0:199:151,40:151:4953:40:908:-16.9953,0,-351.675 0/0:99:985:842,128:842:28502:128:3004:0,-47.9328,-2006.41 0/0:99:512:418,74:418:13832:74:1502:0,-23.3187,-984.518 ./.:.:.:.:.:.:.:.:. 0/0:99:232:193,30:193:6401:30:711:0,-9.69112,-454.514 0/1:0:17:13,4:13:469:4:89:-2.12488,0,-34.6904 0/0:99:874:748,93:748:24700:93:2174:0,-77.1869,-1751.28 0/0:99:712:614,80:614:20430:80:1646:0,-67.1339,-1509.98 0/0:99:816:730,66:730:24806:66:1439:0,-125.684,-1822.58 0/0:99:538:443,82:443:15043:82:1781:0,-9.93394,-1016.46 0/0:99:188:160,24:160:5330:24:389:0,-22.6474,-382.217 0/0:99:563:501,50:501:16369:50:1168:0,-73.0384,-1190.91 0/1:0:45:38,7:38:1318:7:161:-0.919608,0,-93.4117 0/0:99:398:333,57:333:10803:57:1068:0,-35.271,-785.276 0/0:99:218:184,23:184:6088:23:513:0,-24.0412,-440.405 0/0:99:728:660,54:660:22018:54:1111:0,-121.784,-1699.24 0/0:99:355:302,43:302:10302:43:826:0,-31.9642,-720.162 0/1:0:8:5,3:5:161:3:48:-2.00574,0,-11.6892 0/0:99:59:49,8:49:1699:8:175:0,-3.87806,-129.594 0/0:72:35:27,6:27:961:6:123:0,-0.202619,-66.9905 0/0:99:641:543,86:543:18095:86:1899:0,-28.1316,-1274.13 0/0:99:615:539,61:539:17857:61:1307:0,-78.2955,-1339.84 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:.
## NC_007175.2 10968 . T G 0.000292088 . DPB=30;EPPR=4.45795;GTI=0;MQMR=39.5833;NS=1;NUMALT=1;ODDS=25.755;PAIREDR=1;PQR=0;PRO=0;QR=826;RO=24;RPPR=12.0581;SRF=11;SRP=3.37221;SRR=13;DP=10330;AB=0.25;ABP=9.52472;AF=0.5;AO=3;CIGAR=1X;DPRA=0;EPP=9.52472;LEN=1;MEANALT=1;MQM=41.3333;PAIRED=1;PAO=0;PQA=0;QA=47;RPL=0;RPP=9.52472;RPR=3;RUN=1;SAF=0;SAP=9.52472;SAR=3;TYPE=snp;AN=32;AC=2 GT:GQ:DP:AD:RO:QR:AO:QA:GL 0/0:99:30:24,4:24:826:4:53:0,-3.70077,-63.4378 0/0:99:60:51,7:51:1749:7:133:0,-6.40372,-135.949 ./.:.:.:.:.:.:.:.:. 0/0:99:1723:1610,90:1610:56108:90:1320:0,-398.369,-4702.41 0/0:99:1454:1324,114:1324:45488:114:1617:0,-297.358,-3778.2 0/0:99:50:41,8:41:1417:8:102:0,-5.73793,-114.628 0/0:99:568:521,40:521:17979:40:575:0,-117.94,-1496.11 0/0:99:47:39,6:39:1353:6:127:0,-2.30327,-104.561 0/0:99:1933:1783,132:1783:60625:132:2013:0,-411.397,-5054.5 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. 0/0:99:91:71,20:71:2465:20:257:0,-4.50985,-189.426 ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. ./.:.:.:.:.:.:.:.:. 0/0:99:1112:1031,66:1031:35637:66:876:0,-252.949,-2984.74 ./.:.:.:.:.:.:.:.:. 0/0:99:46:41,4:41:1467:4:56:0,-8.6219,-123.072 0/0:99:68:55,12:55:1927:12:212:0,-1.8433,-148.457 0/0:99:1382:1292,72:1292:45198:72:1041:0,-326.041,-3783.22 0/0:99:1701:1571,107:1571:54235:107:1501:0,-375.694,-4546.25 0/1:0:53:41,12:41:1431:12:225:-4.14708,0,-109.171 0/1:0:12:9,3:9:307:3:47:-0.765558,0,-23.4888
### 2.1.1 Trying to get Genetic Relatedness Matrix
ngsDist can also be used to compute genetic distance matrices from
next-generation sequencing (NGS) data. ngsDist is a part of the ngsTools
suite () that provides various
tools for analyzing NGS data.
Convert the VCF file to genotype likelihoods file (GLF) or binary
alignment/map (BAM) format: ngsDist requires input data in GLF or BAM
format. If you have a BAM file, you can use the ‘angsd’ tool from
ngsTools to compute genotype likelihoods. If you have a VCF file, you
can use ‘bcftools’ to convert it to GLF format. Here’s an example of how
to convert a VCF file to GLF format using bcftools:
bcftools query -e 'GT="mis" || GT="hom"' -f '[\%SAMPLE\t%CHROM\t%POS\t%REF\t%ALT\t%GL\n]' input.vcf > input.glf
``` bash
/home/shared/bcftools-1.14/bcftools query \
-e 'GT="mis" || GT="hom"' -f '[%SAMPLE\t%CHROM\t%POS\t%REF\t%ALT\t%GL\n]' ../output/51-SNPs/EpiDiv_merged.f.recode.vcf > ../output/51-SNPs/EpiDiv_merged.f.recode_mishom.glf
```
``` bash
head -2 ../output/51-SNPs/EpiDiv_merged.f.recode_mishom.glf
tail -2 ../output/51-SNPs/EpiDiv_merged.f.recode_mishom.glf
```
## unknown NC_035780.1 451118 A G -110.038,0,-42.8388
## 2:unknown NC_035780.1 451118 A G -122.503,0,-29.1941
## 25:unknown NC_035789.1 19809024 A G -184.391,0,-76.653
## 26:unknown NC_035789.1 19809024 A G -63.4859,0,-117.109
``` bash
/home/shared/bcftools-1.14/bcftools query \
-f'[%CHROM\t%POS\t%REF\t%ALT\t%GL\n]' ../output/51-SNPs/EpiDiv_merged.f.recode.vcf > ../output/51-SNPs/likliehood.txt
```
``` bash
tail ../output/51-SNPs/likliehood.txt
```
## NC_007175.2 10968 T G .
## NC_007175.2 10968 T G .
## NC_007175.2 10968 T G 0,-252.949,-2984.74
## NC_007175.2 10968 T G .
## NC_007175.2 10968 T G 0,-8.6219,-123.072
## NC_007175.2 10968 T G 0,-1.8433,-148.457
## NC_007175.2 10968 T G 0,-326.041,-3783.22
## NC_007175.2 10968 T G 0,-375.694,-4546.25
## NC_007175.2 10968 T G -4.14708,0,-109.171
## NC_007175.2 10968 T G -0.765558,0,-23.4888
``` python
input_file = "../output/51-SNPs/likliehood.txt"
output_file = "../output/51-SNPs/EpiDiv_merged.f.recode.glf"
with open(input_file, "r") as infile, open(output_file, "w") as outfile:
for line in infile:
fields = line.strip().split("\t")
chrom, pos, ref, alt, gl = fields
gl_values = gl.split(",")
outfile.write(f"{chrom}\t{pos}\t{ref}\t{alt}\t{' '.join(gl_values)}\n")
```
``` bash
tail ../output/51-SNPs/EpiDiv_merged.f.recode.glf
```
## NC_007175.2 10968 T G .
## NC_007175.2 10968 T G .
## NC_007175.2 10968 T G 0 -252.949 -2984.74
## NC_007175.2 10968 T G .
## NC_007175.2 10968 T G 0 -8.6219 -123.072
## NC_007175.2 10968 T G 0 -1.8433 -148.457
## NC_007175.2 10968 T G 0 -326.041 -3783.22
## NC_007175.2 10968 T G 0 -375.694 -4546.25
## NC_007175.2 10968 T G -4.14708 0 -109.171
## NC_007175.2 10968 T G -0.765558 0 -23.4888
``` bash
#/home/shared/ngsTools/angsd/angsd \
#-doGlF 2 -doMajorMinor 1 -GL 2 \
#-out ../output/51-SNPs/EpiDiv_merged.f.recode.glf \
#-vcf-pl ../output/51-SNPs/EpiDiv_merged.f.recode.vcf
```
``` bash
head -3 ../output/51-SNPs/EpiDiv_merged.f.recode.glf
tail -3 ../output/51-SNPs/EpiDiv_merged.f.recode.glf
```
## NC_035780.1 123 C T -560.96 0 -28.9657
## NC_035780.1 123 C T -639.833 0 -51.7318
## NC_035780.1 123 C T -334.948 0 -72.3497
## NC_007175.2 10968 T G 0 -375.694 -4546.25
## NC_007175.2 10968 T G -4.14708 0 -109.171
## NC_007175.2 10968 T G -0.765558 0 -23.4888
Create a sites file: ngsDist requires a sites file, which is a
tab-separated file containing information about the sites to be
analyzed. You can create this file from the filtered VCF file:
awk '{print $1"\t"$2}' filtered.recode.vcf > sites.txt
``` bash
awk '{print $1"\t"$2}' ../output/51-SNPs/EpiDiv_merged.f.recode.vcf \
> ../output/51-SNPs/sites.txt
```
``` bash
#tail -1 ../output/51-SNPs/EpiDiv_merged.f.recode.vcf
head -10 ../output/51-SNPs/sites.txt
```
## ##fileformat=VCFv4.2
## ##FILTER=
## ##contig=
## ##contig=
## ##contig=
## ##contig=
``` bash
wc -l ../output/51-SNPs/sites.txt
```
## 2343713 ../output/51-SNPs/sites.txt
4. Run ngsDist: Now, you can run ngsDist using the GLF or BAM file, and
the sites file you created:
``` bash
/home/shared/ngsTools/ngsDist/ngsDist --geno ../output/51-SNPs/EpiDiv_merged.f.recode.glf \ --probs --n_ind 26 \
--n_sites 2343713 --out ../output/51-SNPs/genotype.txt
```
# 3 BS-SNPer
## 3.1 draft paper
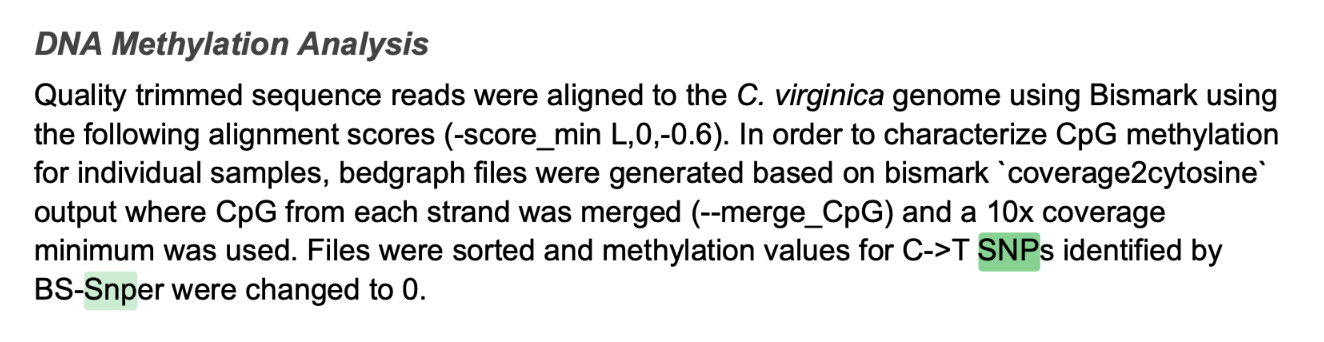
## 3.2 Wiki
10x coverage files (SNP corrected)
at
$$sampleID$$\_R1_val_1\_10x.SNPcorr.bedgraph
## 3.3 issue (backtracking)
see
``` bash
# Directories and programs
bismark_dir="/gscratch/srlab/programs/Bismark-0.22.3/"
bowtie2_dir="/gscratch/srlab/programs/bowtie2-2.3.4.1-linux-x86_64/"
samtools="/gscratch/srlab/programs/samtools-1.9/samtools/"
reads_dir="/gscratch/srlab/sr320/data/cvirg/"
fastqc="/gscratch/srlab/programs/fastqc_v0.11.9/fastqc"
genome_folder="/gscratch/srlab/sr320/data/Cvirg-genome/"
bcftools_dir="/gscratch/srlab/programs/bcftools-1.9/"
htlslib_dir="/gscratch/srlab/programs/htslib-1.9/"
source /gscratch/srlab/programs/scripts/paths.sh
#
# Ah sorry, I always forget about this with bcftools.
#
#Loop through files and compress like this:
#
# bcftools view -Oz -o compressed.vcf.gz plain.vcf
# htsfile compressed.vcf.gz
# bcftools index compressed.vcf.gz
# FILES=$(ls *vcf)
#
# for file in ${FILES}
# do
# NAME=$(echo ${file} | awk -F "." '{print $1}')
# echo ${NAME}
#
# /gscratch/srlab/programs/bcftools-1.9/bcftools view -O z -o ${NAME}.compressed.vcf.gz \
# ${NAME}.SNP-results.vcf
# /gscratch/srlab/programs/htslib-1.9/htsfile ${NAME}.compressed.vcf.gz
# /gscratch/srlab/programs/bcftools-1.9/bcftools index ${NAME}.compressed.vcf.gz
# done
#
#
# /gscratch/srlab/programs/bcftools-1.9/bcftools merge \
# 12M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 44F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 13M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 48M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 16F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 50F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 19F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 52F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 22F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 53F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 23M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 54F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 29F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 59M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 31M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 64M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 35F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 6M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 36F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 76F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 39F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 77F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 3F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 7M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 41F_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# 9M_R1_val_1_bismark_bt2_pe.compressed.vcf.gz \
# --merge all \
# --threads 20 \
# -O v \
# -o Cv_10x_merged.vcf
/gscratch/srlab/programs/vcftools-0.1.16/bin/vcftools \
--vcf Cv_10x_merged.vcf \
--recode --recode-INFO-all \
--min-alleles 2 --max-alleles 2 \
--max-missing 0.5 \
--mac 2 \
--out Cv_10x_merged.filtered
```
Merged VCF -
16G
filtered
file..
``` bash
head ../data/Cv_10x_fxmerge_05.recode.vcf
```
## ##fileformat=VCFv4.2
## ##FILTER=
## ##fileDate= 20220213
## ##bssnperVersion=1.1
## ##bssnperCommand=--fa ../data/Cvirginica_v300.fa --input /home/sr320/github/2018_L18-adult-methylation/bg_data/12M_R1_val_1_bismark_bt2_pe.deduplicated.sorted.bam --output ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.SNP-candidates.txt --methcg ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.CpG-meth-info.tab --methchg ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.CHG-meth-info.tab --methchh ../analyses/bsnp02/12M_R1_val_1_bismark_bt2_pe.CHH-meth-info.tab --minhetfreq 0.1 --minhomfreq 0.85 --minquali 15 --mincover 10 --maxcover 1000 --minread2 2 --errorate 0.02 --mapvalue 20
## ##reference=file://../data/Cvirginica_v300.fa
## ##Bisulfite=directional>
## ##contig=
## ##contig=
## ##contig=
``` bash
tail -2 ../data/Cv_10x_fxmerge_05.recode.vcf
```
## NC_007175.2 12774 . C T 1000 PASS DP=91497;ADF=0,0;ADR=0,220;AD=0,220;AN=52;AC=26 GT:DP:ADF:ADR:AD:BSD:BSQ:ALFR 0/1:407:0,0:0,407:0,407:0,0,223,0,0,407,0,0:0,0,36,0,0,37,0,0:0,1 0/1:5786:0,0:8,5778:8,5778:0,0,4267,1,3,5778,8,0:0,0,36,25,37,36,36,0:0.001,0.999 0/1:557:0,0:1,556:1,556:0,0,391,0,1,556,1,0:0,0,36,0,37,37,37,0:0.002,0.998 0/1:819:0,0:0,819:0,819:0,0,695,0,0,819,0,0:0,0,36,0,0,36,0,0:0,1 0/1:2838:0,0:6,2832:6,2832:0,0,1829,0,1,2832,6,0:0,0,36,0,37,36,35,0:0.002,0.998 0/1:4256:0,0:9,4247:9,4247:0,0,2781,0,4,4247,9,1:0,0,36,0,34,36,37,37:0.002,0.998 0/1:5216:0,0:9,5207:9,5207:0,0,6064,0,2,5207,9,1:0,0,36,0,31,36,37,37:0.002,0.998 0/1:3184:0,0:2,3182:2,3182:0,0,1748,0,1,3182,2,1:0,0,36,0,37,36,37,25:0.001,0.999 0/1:5953:0,0:7,5946:7,5946:0,0,3806,0,0,5946,7,1:0,0,36,0,0,37,35,25:0.001,0.999 0/1:7521:0,0:10,7511:10,7511:0,0,5104,5,7,7511,10,1:0,0,36,25,35,36,36,25:0.001,0.999 0/1:386:0,0:2,384:2,384:0,0,299,0,0,384,2,0:0,0,36,0,0,37,37,0:0.005,0.995 0/1:4995:0,0:8,4987:8,4987:0,0,3422,1,1,4987,8,1:0,0,36,25,37,36,37,37:0.002,0.998 0/1:2953:0,0:2,2951:2,2951:0,0,2036,0,1,2951,2,0:0,0,36,0,37,36,37,0:0.001,0.999 0/1:288:0,0:2,286:2,286:0,0,167,0,0,286,2,0:0,0,36,0,0,36,37,0:0.007,0.993 0/1:283:0,0:4,279:4,279:0,0,195,0,0,279,4,0:0,0,36,0,0,36,37,0:0.014,0.986 0/1:747:0,0:3,744:3,744:0,0,554,0,0,744,3,0:0,0,36,0,0,37,37,0:0.004,0.996 0/1:8439:0,0:17,8422:17,8422:0,0,6163,3,5,8422,17,0:0,0,36,37,37,36,36,0:0.002,0.998 0/1:597:0,0:0,597:0,597:0,0,494,2,3,597,0,0:0,0,36,25,37,36,0,0:0,1 0/1:6748:0,0:8,6740:8,6740:0,0,5233,1,6,6740,8,0:0,0,36,25,37,36,37,0:0.001,0.999 0/1:6916:0,0:8,6908:8,6908:0,0,6197,1,2,6908,8,0:0,0,36,25,37,37,37,0:0.001,0.999 0/1:7641:0,0:11,7630:11,7630:0,0,6446,2,5,7630,11,0:0,0,36,25,37,36,36,0:0.001,0.999 0/1:7115:0,0:4,7111:4,7111:1,0,6214,1,4,7111,4,0:25,0,36,25,37,36,37,0:0.001,0.999 0/1:5198:0,0:3,5195:3,5195:0,0,4280,0,1,5195,3,1:0,0,36,0,37,36,37,37:0.001,0.999 0/1:521:0,0:2,519:2,519:0,0,287,0,0,519,2,0:0,0,36,0,0,36,37,0:0.004,0.996 0/1:1913:0,0:4,1909:4,1909:0,0,1107,0,0,1909,4,2:0,0,36,0,0,37,37,37:0.002,0.998 0/1:220:0,0:0,220:0,220:0,0,158,0,0,220,0,0:0,0,36,0,0,37,0,0:0,1
## NC_007175.2 15247 . C T 1000 PASS DP=43802;ADF=0,0;ADR=0,57;AD=0,57;AN=52;AC=26 GT:DP:ADF:ADR:AD:BSD:BSQ:ALFR 0/1:112:0,0:0,112:0,112:0,0,322,0,0,112,0,0:0,0,36,0,0,36,0,0:0,1 0/1:2287:0,0:5,2282:5,2282:0,0,3893,1,2,2282,5,1:0,0,36,25,37,36,35,25:0.002,0.998 0/1:174:0,0:0,174:0,174:0,0,408,0,0,174,0,0:0,0,37,0,0,36,0,0:0,1 0/1:312:0,0:0,312:0,312:0,0,805,0,0,312,0,0:0,0,36,0,0,36,0,0:0,1 0/1:1310:0,0:0,1310:0,1310:0,0,1883,0,0,1310,0,0:0,0,36,0,0,36,0,0:0,1 0/1:2014:0,0:8,2006:8,2006:0,0,2965,0,3,2006,8,0:0,0,36,0,33,36,36,0:0.004,0.996 0/1:4017:0,0:7,4010:7,4010:0,0,6081,0,6,4010,7,0:0,0,36,0,37,36,37,0:0.002,0.998 0/1:1398:0,0:2,1396:2,1396:0,0,1817,0,2,1396,2,0:0,0,36,0,37,36,37,0:0.001,0.999 0/1:2263:0,0:2,2261:2,2261:0,0,3922,1,3,2261,2,0:0,0,36,25,33,36,37,0:0.001,0.999 0/1:3506:0,0:2,3504:2,3504:0,0,4504,0,3,3504,2,0:0,0,36,0,37,36,37,0:0.001,0.999 0/1:140:0,0:2,138:2,138:0,0,291,0,0,138,2,0:0,0,36,0,0,36,37,0:0.014,0.986 0/1:2187:0,0:0,2187:0,2187:0,0,3267,0,0,2187,0,1:0,0,36,0,0,36,0,25:0,1 0/1:1099:0,0:3,1096:3,1096:1,0,1999,0,0,1096,3,0:37,0,36,0,0,36,29,0:0.003,0.997 0/1:106:0,0:0,106:0,106:0,0,182,0,0,106,0,0:0,0,36,0,0,36,0,0:0,1 0/1:136:0,0:0,136:0,136:0,0,249,0,0,136,0,0:0,0,36,0,0,36,0,0:0,1 0/1:230:0,0:0,230:0,230:0,0,540,0,0,230,0,0:0,0,36,0,0,36,0,0:0,1 0/1:4059:0,0:9,4050:9,4050:0,0,5635,1,5,4050,9,2:0,0,36,37,37,36,36,31:0.002,0.998 0/1:188:0,0:0,188:0,188:0,0,624,1,0,188,0,0:0,0,36,25,0,36,0,0:0,1 0/1:3105:0,0:3,3102:3,3102:1,0,4920,0,4,3102,3,1:25,0,36,0,34,36,37,25:0.001,0.999 0/1:3066:0,0:3,3063:3,3063:0,0,6416,2,3,3063,3,0:0,0,36,37,33,36,37,0:0.001,0.999 0/1:4533:0,0:6,4527:6,4527:0,0,5452,1,2,4527,6,0:0,0,36,37,37,36,35,0:0.001,0.999 0/1:3451:0,0:5,3446:5,3446:0,0,6195,2,1,3446,5,2:0,0,36,25,37,36,37,37:0.001,0.999 0/1:2841:0,0:0,2841:0,2841:1,0,4298,0,3,2841,0,1:25,0,36,0,33,36,0,25:0,1 0/1:159:0,0:0,159:0,159:0,0,373,0,0,159,0,0:0,0,36,0,0,36,0,0:0,1 0/1:1052:0,0:4,1048:4,1048:0,0,1293,1,0,1048,4,0:0,0,36,25,0,36,37,0:0.004,0.996 0/1:57:0,0:0,57:0,57:0,0,165,0,0,57,0,0:0,0,37,0,0,35,0,0:0,1