cd ../data
curl -O https://gannet.fish.washington.edu/seashell/snaps/PGA_assembly.scaffolds_only.gff3Evaluating Assemblies
Rpubs link
https://rpubs.com/sr320/1042307
Check out which assembly has best gene set
https://github.com/laurahspencer/DuMOAR/issues/25
GAWN (Genome Annotation Without Nightmares):
Following the GAWN instructions I ran the script GAWN-annotation.sh using the scaffold-only genome, the transcriptome that Giles generated for me, the Swissprot database, and the config file gawn_config.sh. All resulting files are on Google Drive here, but the files <100MB are also in this repo here. Feel free to drop any files resulting from your work in either location.
Gawn: https://gannet.fish.washington.edu/seashell/snaps/PGA_assembly.scaffolds_only.gff3
GenSaS:
All resulting files are on Google Drive here, and files <100 MB are also in this repo here (which includes all except for the repeat elements). If anyone has a GenSaS account I can share the project with you, I just need to know your username. Note that GenSaS was run with the full genome, so any resulting files should be filtered for only the scaffolds.
GenSas
https://gannet.fish.washington.edu/seashell/snaps/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.gff3
https://gannet.fish.washington.edu/seashell/snaps/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.fna
GAWN
head ../data/PGA_assembly.scaffolds_only.gff3##gff-version 3
# Generated by GMAP version 2021-08-25 using call: gmap.sse42 -t 20 --dir 03_data -d indexed_genome -f gff3_gene --gff3-add-separators=0
PGA_scaffold1__111_contigs__length_23635802 indexed_genome gene 6977635 6977908 . + . ID=TRINITY_DN8_c4_g2_i1.path1;Name=TRINITY_DN8_c4_g2_i1;Dir=indeterminate
PGA_scaffold1__111_contigs__length_23635802 indexed_genome mRNA 6977635 6977908 . + . ID=TRINITY_DN8_c4_g2_i1.mrna1;Name=TRINITY_DN8_c4_g2_i1;Parent=TRINITY_DN8_c4_g2_i1.path1;Dir=indeterminate;coverage=97.9;identity=98.5;matches=270;mismatches=4;indels=0;unknowns=0
PGA_scaffold1__111_contigs__length_23635802 indexed_genome exon 6977635 6977908 98 + . ID=TRINITY_DN8_c4_g2_i1.mrna1.exon1;Name=TRINITY_DN8_c4_g2_i1;Parent=TRINITY_DN8_c4_g2_i1.mrna1;Target=TRINITY_DN8_c4_g2_i1 274 1 .
PGA_scaffold1__111_contigs__length_23635802 indexed_genome CDS 6977636 6977767 99 + 0 ID=TRINITY_DN8_c4_g2_i1.mrna1.cds1;Name=TRINITY_DN8_c4_g2_i1;Parent=TRINITY_DN8_c4_g2_i1.mrna1;Target=TRINITY_DN8_c4_g2_i1 133 2 .
PGA_scaffold15__74_contigs__length_14466968 indexed_genome gene 2954382 2964582 . + . ID=TRINITY_DN8_c5_g1_i11.path1;Name=TRINITY_DN8_c5_g1_i11;Dir=antisense
PGA_scaffold15__74_contigs__length_14466968 indexed_genome mRNA 2954382 2964582 . + . ID=TRINITY_DN8_c5_g1_i11.mrna1;Name=TRINITY_DN8_c5_g1_i11;Parent=TRINITY_DN8_c5_g1_i11.path1;Dir=antisense;coverage=100.0;identity=99.5;matches=402;mismatches=2;indels=0;unknowns=0
PGA_scaffold15__74_contigs__length_14466968 indexed_genome exon 2954382 2954466 100 + . ID=TRINITY_DN8_c5_g1_i11.mrna1.exon1;Name=TRINITY_DN8_c5_g1_i11;Parent=TRINITY_DN8_c5_g1_i11.mrna1;Target=TRINITY_DN8_c5_g1_i11 320 404 -
PGA_scaffold15__74_contigs__length_14466968 indexed_genome exon 2964264 2964582 99 + . ID=TRINITY_DN8_c5_g1_i11.mrna1.exon2;Name=TRINITY_DN8_c5_g1_i11;Parent=TRINITY_DN8_c5_g1_i11.mrna1;Target=TRINITY_DN8_c5_g1_i11 1 319 -awk '$3=="gene"' ../data/PGA_assembly.scaffolds_only.gff3 > ../results/PGA_assembly.scaffolds_only.gene.gff3tail ../results/PGA_assembly.scaffolds_only.gene.gff3PGA_scaffold28__73_contigs__length_10794191 indexed_genome gene 1938804 1952116 . - . ID=TRINITY_DN182802_c0_g1_i1.path2;Name=TRINITY_DN182802_c0_g1_i1;Dir=indeterminate
PGA_scaffold7__77_contigs__length_16129408 indexed_genome gene 15030732 15030773 . + . ID=TRINITY_DN182802_c0_g1_i1.path3;Name=TRINITY_DN182802_c0_g1_i1;Dir=indeterminate
PGA_scaffold7__77_contigs__length_16129408 indexed_genome gene 15029337 15029374 . + . ID=TRINITY_DN182802_c0_g1_i1.path4;Name=TRINITY_DN182802_c0_g1_i1;Dir=indeterminate
PGA_scaffold1__111_contigs__length_23635802 indexed_genome gene 2355847 2355883 . - . ID=TRINITY_DN182802_c0_g1_i1.path5;Name=TRINITY_DN182802_c0_g1_i1;Dir=indeterminate
PGA_scaffold2__216_contigs__length_42616187 indexed_genome gene 37241209 37241287 . + . ID=TRINITY_DN161898_c0_g1_i1.path1;Name=TRINITY_DN161898_c0_g1_i1;Dir=indeterminate
PGA_scaffold46__73_contigs__length_12693875 indexed_genome gene 3575215 3575292 . + . ID=TRINITY_DN161898_c0_g1_i1.path2;Name=TRINITY_DN161898_c0_g1_i1;Dir=indeterminate
PGA_scaffold46__73_contigs__length_12693875 indexed_genome gene 3555535 3555601 . + . ID=TRINITY_DN161898_c0_g1_i1.path3;Name=TRINITY_DN161898_c0_g1_i1;Dir=indeterminate
PGA_scaffold43__161_contigs__length_26349757 indexed_genome gene 4053856 4053947 . - . ID=TRINITY_DN161898_c0_g1_i1.path4;Name=TRINITY_DN161898_c0_g1_i1;Dir=indeterminate
PGA_scaffold43__161_contigs__length_26349757 indexed_genome gene 4054015 4054068 . - . ID=TRINITY_DN161898_c0_g1_i1.path5;Name=TRINITY_DN161898_c0_g1_i1;Dir=indeterminate
PGA_scaffold34__18_contigs__length_2048619 indexed_genome gene 681313 724568 . + . ID=TRINITY_DN183092_c0_g1_i1.path1;Name=TRINITY_DN183092_c0_g1_i1;Dir=indeterminatewc -l ../results/PGA_assembly.scaffolds_only.gene.gff3185015 ../results/PGA_assembly.scaffolds_only.gene.gff3grab fasta
/home/shared/bedtools2/bin/bedtools getfasta \
-fi ../data/Mmag_scaffold.fa \
-bed ../results/PGA_assembly.scaffolds_only.gene.gff3 \
-fo ../results/GAWN-gene.fa/home/shared/bedtools2/bin/bedtools getfasta \
-fi ../data/Mmag_scaffold.fa \
-bed ../results/PGA_assembly.scaffolds_only.gene.gff3 \
| tail# Read in FastA file
fasta_filegawn <- "../results/GAWN-gene.fa"
sequences <- readDNAStringSet(fasta_filegawn)
# Calculate lengths
lengths <- width(sequences)
# Create a data frame
df <- data.frame(length = lengths)
# Plot histogram with ggplot2
ggplot(df, aes(x = length)) +
geom_histogram(binwidth = 1000, fill = "blue", color = "black") +
xlab("Sequence Length") +
ylab("Frequency") +
ggtitle("Histogram of Sequence Lengths") +
theme_minimal()GenSas
https://gannet.fish.washington.edu/seashell/snaps/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.gff3
https://gannet.fish.washington.edu/seashell/snaps/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.fna
cd ../data
curl -O https://gannet.fish.washington.edu/seashell/snaps/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.gff3
curl -O https://gannet.fish.washington.edu/seashell/snaps/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.fnahead ../data/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.gff3##gff-version 3
##Generated using GenSAS, Tuesday 25th of April 2023 11:25:49 AM
##Project Name : Dungeness crab genome annotation
##Job Name : Annotations a1
##Tool : Publish
PGA_scaffold0__40_contigs__length_4818635 GenSAS_6448195bde6ca-publish gene 309 602 . + . ID=M_mag.00g000010-v1.0.a1;Name=M_mag.00g000010;
PGA_scaffold0__40_contigs__length_4818635 GenSAS_6448195bde6ca-publish mRNA 309 602 588.00 + . ID=M_mag.00g000010.m01-v1.0.a1;Name=M_mag.00g000010.m01;Parent=M_mag.00g000010-v1.0.a1;
PGA_scaffold0__40_contigs__length_4818635 GenSAS_6448195bde6ca-publish exon 309 602 . + . ID=M_mag.00g000010.m01.exon01-v1.0.a1;Name=M_mag.00g000010.m01.exon01;Parent=M_mag.00g000010.m01-v1.0.a1;
PGA_scaffold0__40_contigs__length_4818635 GenSAS_6448195bde6ca-publish CDS 309 602 . + 0 ID=M_mag.00g000010.m01.CDS01-v1.0.a1;Name=M_mag.00g000010.m01.CDS01;Parent=M_mag.00g000010.m01-v1.0.a1;
PGA_scaffold0__40_contigs__length_4818635 GenSAS_6448195bde6ca-publish gene 3038 3340 . + . ID=M_mag.00g000020-v1.0.a1;Name=M_mag.00g000020;awk '!/unscaffolded/' ../data/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.gff3 > ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes-scaffold.gff3awk '$3=="gene"' ../data/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.gff3 | awk '!/unscaffolded/' > ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.gene-only.gff3tail ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.gene-only.gff3PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14019156 14020396 . + . ID=M_mag.00g437910-v1.0.a1;Name=M_mag.00g437910;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14043020 14043864 . - . ID=M_mag.00g437920-v1.0.a1;Name=M_mag.00g437920;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14046610 14047137 . - . ID=M_mag.00g437930-v1.0.a1;Name=M_mag.00g437930;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14066890 14069984 . - . ID=M_mag.00g437940-v1.0.a1;Name=M_mag.00g437940;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14076331 14076585 . + . ID=M_mag.00g437950-v1.0.a1;Name=M_mag.00g437950;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14077692 14078243 . - . ID=M_mag.00g437960-v1.0.a1;Name=M_mag.00g437960;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14079232 14081077 . - . ID=M_mag.00g437970-v1.0.a1;Name=M_mag.00g437970;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14081929 14082627 . - . ID=M_mag.00g437980-v1.0.a1;Name=M_mag.00g437980;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14102371 14102958 . - . ID=M_mag.00g437990-v1.0.a1;Name=M_mag.00g437990;
PGA_scaffold48__117_contigs__length_14149252 GenSAS_6448195bde6ca-publish gene 14108083 14108473 . - . ID=M_mag.00g438000-v1.0.a1;Name=M_mag.00g438000;wc -l ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.gene-only.gff343800 ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.gene-only.gff3grep -c ">" ../data/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.fna50294tail ../data/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.genes.fnaAAAGCAAGGCGCCGCAGCGGACCAGCACACGCCAGCCTCCAGGTGGTTCT
GGCAGACTGGTTTTTCATTATCTAG
>M_mag.00g502940-v1.0.a1 ID=M_mag.00g502940-v1.0.a1|Name=M_mag.00g502940|organism=Metacarcinus magister|type=gene|length=309|location=scaffold_75_pilon__unscaffolded:158308..158616+
ATGAAGGAAGATAACAGGGCACACGGAACGGAACAGGACGTCGTGAAAAC
ACACATAGAGAACGGGAAGGCACGTACGCACGAACGAAGACAGGAAAAGA
AGGAAAGAAGAAAGAAAAATCCACGCAAGAAGAGAGTGGAAGATTTGACG
GAAAAAAACAAGGTGGGAAAAACATATAGGAAAAATTATCACAAAAAGAG
GTTGATGGCAGGATGGAGTGGTGTGAGGAGAGACTTAAGGGGGAAGAAAG
GAGAAGCCAAAGAGGAGGAGGAGGAGGAGGAGGAGGAGGAGGTGAAAGTT
AAAGCGTAGgrab fasta
/home/shared/bedtools2/bin/bedtools getfasta \
-fi ../data/Mmag_scaffold.fa \
-bed ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.gene-only.gff3 \
-fo ../results/GenSAS-gene.fa/home/shared/bedtools2/bin/bedtools getfasta \
-fi ../data/Mmag_scaffold.fa \
-bed ../results/Metacarcinus-magister-v1.0.a1.6448195bde6ca-publish.gene-only.gff3 \
| tail>PGA_scaffold48__117_contigs__length_14149252:14077691-14078243
TTAGACATTTGCCTTTGTTTTGGTGTGGGAGATGGTGAAGCCGATTTTCTCACACTTGGCGGCCAGAAGGTTGAGAGAGTCTTGCATTGTGGTGGTGGAGTCTCTGCCTGTTGTTTGTAAGATAATATCGTCAGCATACCCGATGTGTTGTGTCCCTTGTGGATAGGGGAGTTTGGCAAGGGCGTCCATGAGAATGTTAAACAGAGTTGGACTGATGACGCCTCCCTGTGGTGTTCCGTTTTCTAGTTGCATGTAGTTAGACGCCTCCCCCTGGAACACAACACGAGCCTTACTGTTCGTCAGGTAGTGCCGCACCCACGATAGTAATTTCCCTCTCACCCCCATCTTGGTAAGTTCATAGAGGATGGTCAGGGGCTGAGCTCTATCGAAGGCTTTTTCTATGTCGAGGAACACTGCTGTTTTGGCTTGGTGATAAGAATAGTAGTTGGAGAGGCAGTTAACAGTGCTCCTGTGTTTGACAAAACCATTCACACCAACATATAGGCGTCCGATCTTATAGTTTAACCGATTCAGCAGTGTTCTTTCCAGCAT
>PGA_scaffold48__117_contigs__length_14149252:14079231-14081077
CTACACCCCTTGGTGAGGCATGGAGGCATCAGCTGCAGCTTGAATAGCTTGGGTCAGGTCTTCAGCAAAGAGTTGCACCGAGTTAGGGACAGCATAAGTGGCATACCAGGTGTTTAAGTGGCCTGTGAATTTCGTCCAGTCAGCTTTCCTTGTTAGCCATCTTTTCTTTGTGATGTTAGGCTGGTCTGTAGTCCTCGGGAGAGTTATGTTATACTCCTGTGCCCAGTGATCACTTAGTAGTTCAGGTATGCTGTGGACTTCCTTGACACAATCACTGCCATTCAGGGTGAGATCAAGCCTCCCGCCCCCAATATGAGTGACGGATGACGCACCATGCAGTTGTAGTTCCTCGAAGTCCTCCAGCATTTGAACGATGTGACGCCCTCGTAAGTTGCCACCACCTTCACTCCAAGGCTCCAAGAGTGGATGGTGAGCATTGAAGTCTCCACTCAGGACGAGTAGTCCACCTGTATTGCTTGTCGCCGTGTTTAGCAGATCGAGGTTCGACCTTTGTCTGCAATATACGTTACAGACATCCACTTTGACGGAAGTGTCAAAGTGTATCGTGACACAGAGTGAGTCTACCTGTTGTCCGAGATACGGTGGAGTCTGTGTTAGGGTGGCTGAAATATCCCTCCTGACAAGGGTGATGAGCCCCCGAGAGCCTCCTTGCACGTAGGGTTGCCGCATAGCAATTTGCGATGCACACATTATTTTTAGATCCTGGCACTCCTGCATTTTTGTTCGTTGATCTCTGAATGCCTTCAATACCTGCCGTAAAGCTTCTACTTCCCTGAGTAGTTGAGTACTGGATCTCTGGCAGAGTTGCTTGTCTGACTTACTGGTGGTCTCTCTGTAGCAGTTGGATCGGGGGTGGGGTCCTGAAGTACACATTGTTGGGCTTGTTGTTGGGACTGATTGCCAGGTTGTGGGGCTCCTTGTGGCTGTGTTGGCGATGGTTGGTGGCGAGTCCAGGGGTAATGGTTTACCGTGGGGTGGTGGTGGGTGGTGGGTGGTAAGGGTTGACTGGATGTGCTTGGTGAGGTATCGACTGTTAGTGGGTCGGTGGTGAGGGTCGCTGATGGGTCTCCTGAGGCAGGAGTGGAAACGCGCTGGAGGGAGGTACAGGAGCCCCTGCAGGTGTTACCTGACCCTTGGCGGCATGGAGGTGCAAATCTGGTCGTAGTGGACACCTGAAACTCCAAGCGTTATGTTCCTGATAACAGTTTGGTCACTTCGGTGGTACTTTTTCCCCACTGTCTATTCTTTCCCTGCATAGCTTGGGTCGTGATTCCCCGCACAGAATCCGTACCTTGTTTTCCCGTCGCATTGCCACACTCTGTGTCCCCATTTCTGGCAATTGCTACAAAAAGCTGGTTTCCCCACATACTTCTCTGCTTTGCAGGGTCTCATTCCCGGAAACTGCAGTGTTGTTGGTGGTTCTCCCTCCCATAGTGCAAGCACCTGATTCCTTGGTGCCCGGTTGTCGCGGCGTGTCTCTCTCTCCATCCACAATACAGATAGGAGGTCCTCTATGTAGTCTAGTGGCATGTATGATGGATAGTGGCGTATGATTATCTGCATCTGCCTGTTGTCCTAGGTAATCCTCGTCAGCTTCATGTTGTTGTTTTGGTAAGGATGTCCTATCTCGGTGAGGAGCTTGACGGTGTCCATGTCCCGAATCACTGCAATTATGGATTTTGCTGTCATCCTTGGCTGCACTGATAAGTTGCGGTGCTTGTTGGCGACATCCGCCAGCTATATCGCCCTGTCCCTCGGAGGTACGCCCTTCGGGAAGAACAGGCGGTATGTTTCCTGTGCTGGCACAGGATTGGGATGCTAACAT
>PGA_scaffold48__117_contigs__length_14149252:14081928-14082627
CTAGCGGAGGAACCAGGAATCAGAGGTAGCAGTGTGTGGCAACTGTGGTGGTGAAGCACGTGGGGGTGCAGGAGGTGCGGCAGGAGGATGTGAAGGTGTGGGATCAACGCTGCGTCTGCCACACCCATGCCACACGCTCACACCGACCAAGTGACATCGCATATACTTCCAGTGGGAGGTATCGGGTGAGACGGGAGGCTCCGTGGAAGAGGAAGAGGAAGGAGGGGGCGGCGGGGGTTGGCGGTGAATACAGGTGCGCGAATCGTGTGGGCCAACACACCACGCACACTTCTCCGCTGAGCAGTAGCGCGAGATATGACCAATACCCCAGCATTTGAAACACCAGGGTTGGTCATCCTTAATCCTGCGAAGCTCACAGTCTGGGAGATAGGAGAGAAAAGTGAAGGCAACAATAGGTGGGGGCGGCTCCAGCAGGCTCCAGGTGACAACGATGCTGTTGATGGGCACACCGTCCTGACGAAAACGTCGCGCAGAGTAAACAACCGGGAATTCCTTACTGAGGGATGGATCAACATCGACAGGGTAGCGAGTAACCAGGTAGCTGTGATATTTTCTGGACCTTTCCGGTGAGTCCTGAACTTCAAGACTCAGAGACAAAAACTCCCCCTGCATGGACCGTTCCACGATGTCTTCTCGACGCCTGAAGACATACACAAAACGCGATGTGACTGCAGCCAT
>PGA_scaffold48__117_contigs__length_14149252:14102370-14102958
CTAAGGGCGCTGGCGCTTGCGCTCCTCAGCCGCTCGTGCGGCTGCGCCAGTGGCATAAGCACAGTCAAGGAGATGGGTGTCCGCGAATGTGTTGACGCATGTGGCGTCCCACATCAGCATCCTGCCTCTCCTAAAGGGGAAGACTGTGATGCCGTCTGGTCGGCGTCCGTCCCCTCGGTCCAAACCTTGTGGCTCGAGGATGGCTACCAAGCCAGCCGCGGCCAGGGCCCGGTAAACCAAGTCGTTGAGAGCTGCGTGGCGGGGCAGGCGGCCGGGATTCCGGGGGCAGGAGAGGCTGTGGTAGCCGAATGCGTCTGTTATGACTCCACAGCGGCAGCGATGAGGTTGCTGGACGCGTGTGCCCAGCCTCAGTGCGACGCCCACACGGACCGCTTCATTCGGGAGGAGGAGGCCAAGGCACTCTACGGGGATGGCTGACATCCAAGCCCCGCTGTGCGGTGCTGTGGCGGCGAGGAGACGTGCTCGGTCAATCTGGTTCGCGTGAGCCAGCAGGTTGTCCAGACGGTGCTTGGAGGCTAACTCGTCCAGTGTCCGTTGCGTGATGGCGAGATCAGGGCTGAGGTTAGG
>PGA_scaffold48__117_contigs__length_14149252:14108082-14108473
AGGGTATACTACTACTACTACTACTACTACTACTACTACCACCACCACTATTACTACCACTACTACCACCACCACTACTACTACCACTACTATTACTACTACTACCACTACTATTACTATTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACCACTATTACTACCACTACTACCTCCATCACTACTACTACCACTACTATTACTACTATTGCCACTACAACTTTACAACCTACTACTACTACTACTACTACTACTATTATACTATTATTATTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTACTATTATTACTTCTACTATTACTGCTGTTACTA# Read in FastA file
fasta_filegen <- "../results/GenSAS-gene.fa"
sequences <- readDNAStringSet(fasta_filegen)
# Calculate lengths
lengths <- width(sequences)
# Create a data frame
df <- data.frame(length = lengths)
# Plot histogram with ggplot2
ggplot(df, aes(x = length)) +
geom_histogram(binwidth = 1000, fill = "blue", color = "black") +
xlab("Sequence Length") +
ylab("Frequency") +
ggtitle("Histogram of Sequence Lengths") +
theme_minimal()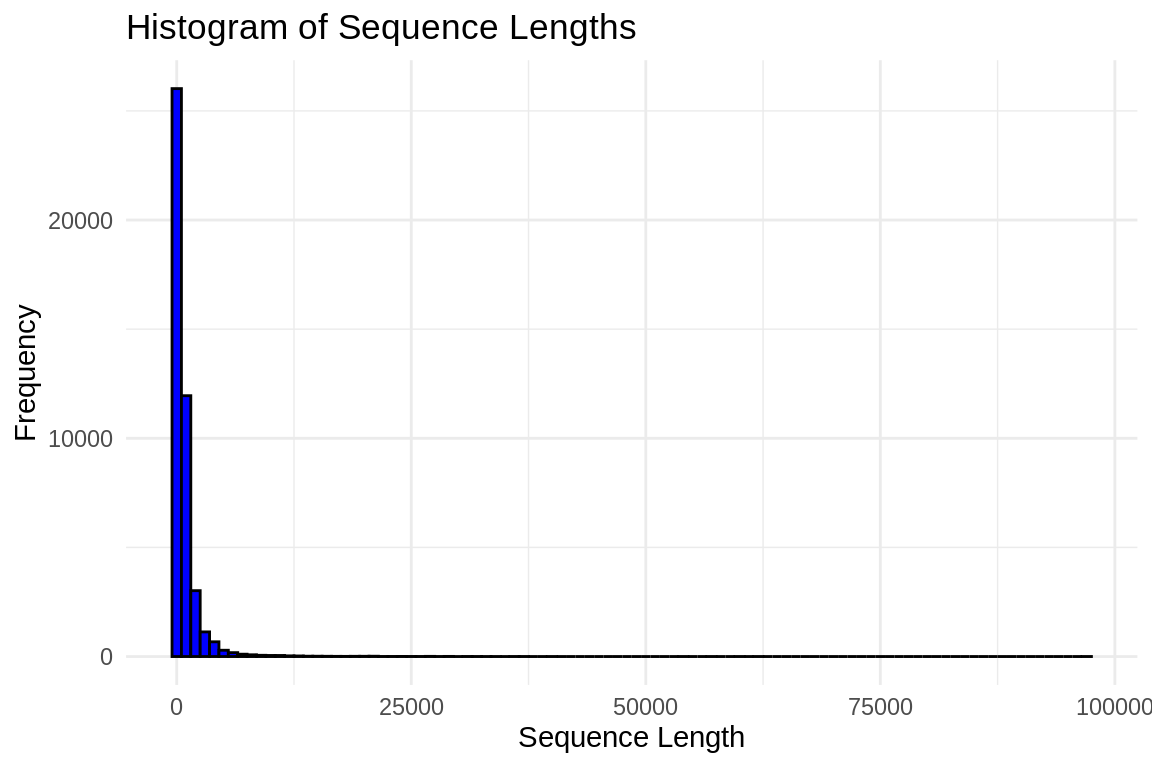
Genome
md5sum ../data/Mmag_scaffold.fa6ad168f282e81c317fff794e1045ed39 ../data/Mmag_scaffold.fagrep '>' ../data/Mmag_scaffold.fa | wc -l49grep '>' ../data/Mmag_scaffold.fa | head>PGA_scaffold0__40_contigs__length_4818635
>PGA_scaffold1__111_contigs__length_23635802
>PGA_scaffold2__216_contigs__length_42616187
>PGA_scaffold3__77_contigs__length_22140449
>PGA_scaffold4__118_contigs__length_24133938
>PGA_scaffold5__54_contigs__length_17529967
>PGA_scaffold6__2_contigs__length_4500
>PGA_scaffold7__77_contigs__length_16129408
>PGA_scaffold8__70_contigs__length_6254265
>PGA_scaffold9__2_contigs__length_6774/home/shared/samtools-1.12/samtools faidx ../data/Mmag_scaffold.fa