Input data and parameters
QualiMap command line
| qualimap bamqc -bam EF07-EM01-Zygote.deduplicated.sorted.bam -nw 400 -hm 3 |
Alignment
| Command line: | "bismark -1 EF07-EM01-Zygote_1_val_1.fq.gz -2 EF07-EM01-Zygote_2_val_2.fq.gz --genome BismarkIndex --bam --bowtie2 --multicore 5" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | yes |
| Program: | Bismark (v0.24.2) |
| Analysis date: | Thu Dec 05 15:48:30 GMT 2024 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | EF07-EM01-Zygote.deduplicated.sorted.bam |
Summary
Globals
| Reference size | 684,741,128 |
| Number of reads | 5,632,404 |
| Mapped reads | 5,632,404 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 5,632,404 / 100% |
| Mapped reads, first in pair | 2,816,202 / 50% |
| Mapped reads, second in pair | 2,816,202 / 50% |
| Mapped reads, both in pair | 5,632,404 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Secondary alignments | 0 |
| Read min/max/mean length | 20 / 150 / 86.4 |
| Overlapping read pairs | 2,803,367 / 99.54% |
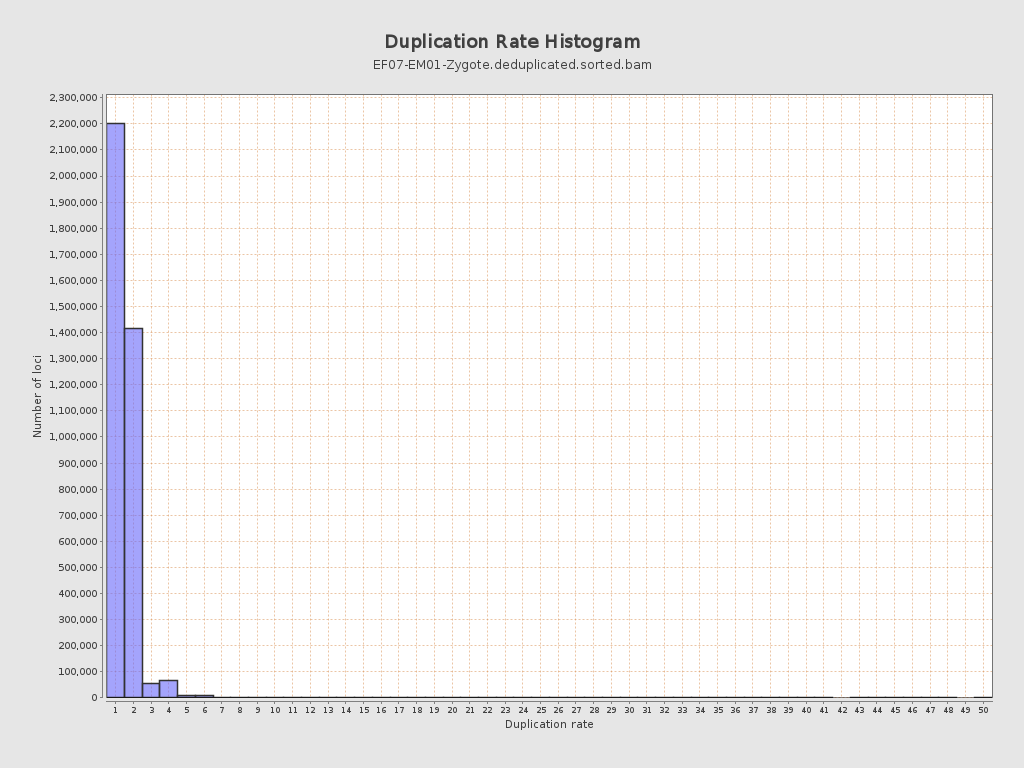
| Duplicated reads (estimated) | 1,871,358 / 33.22% |
| Duplication rate | 41.49% |
| Clipped reads | 0 / 0% |
ACGT Content
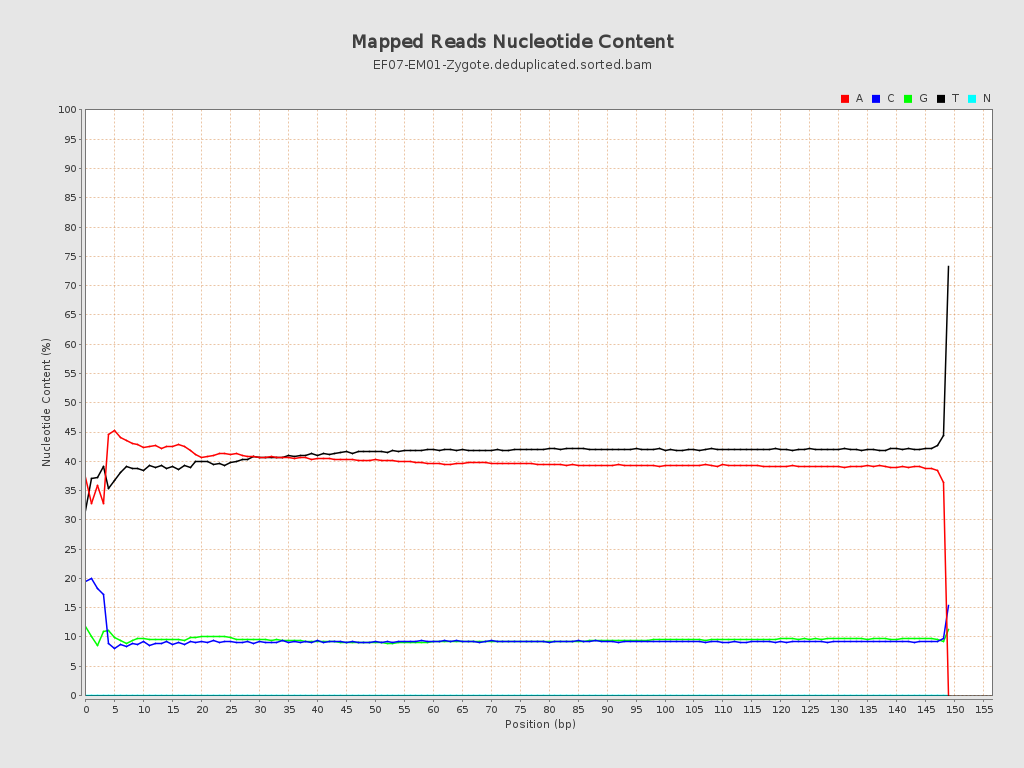
| Number/percentage of A's | 195,796,561 / 40.28% |
| Number/percentage of C's | 46,552,161 / 9.58% |
| Number/percentage of T's | 197,713,007 / 40.68% |
| Number/percentage of G's | 46,007,125 / 9.47% |
| Number/percentage of N's | 2,811 / 0% |
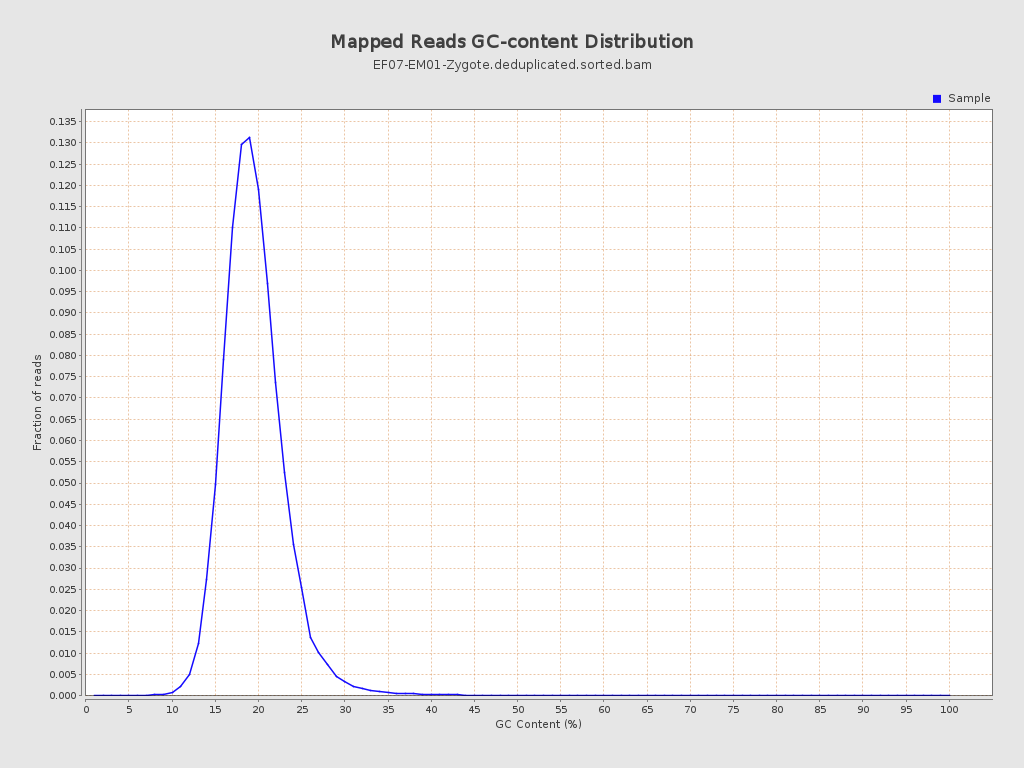
| GC Percentage | 19.04% |
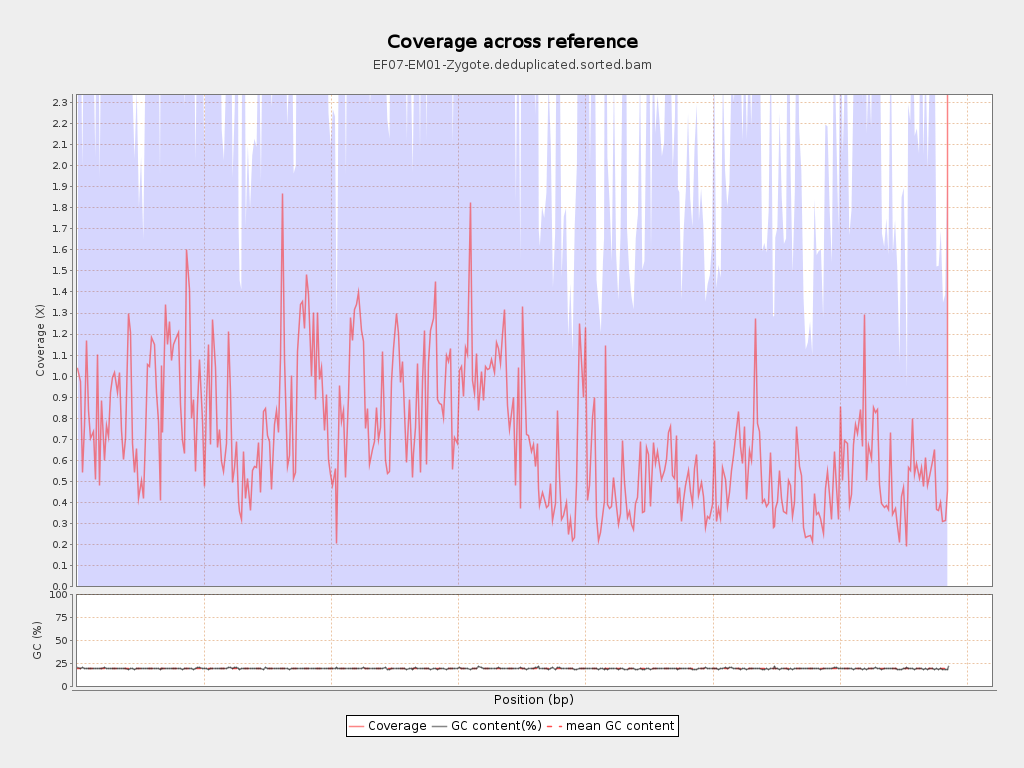
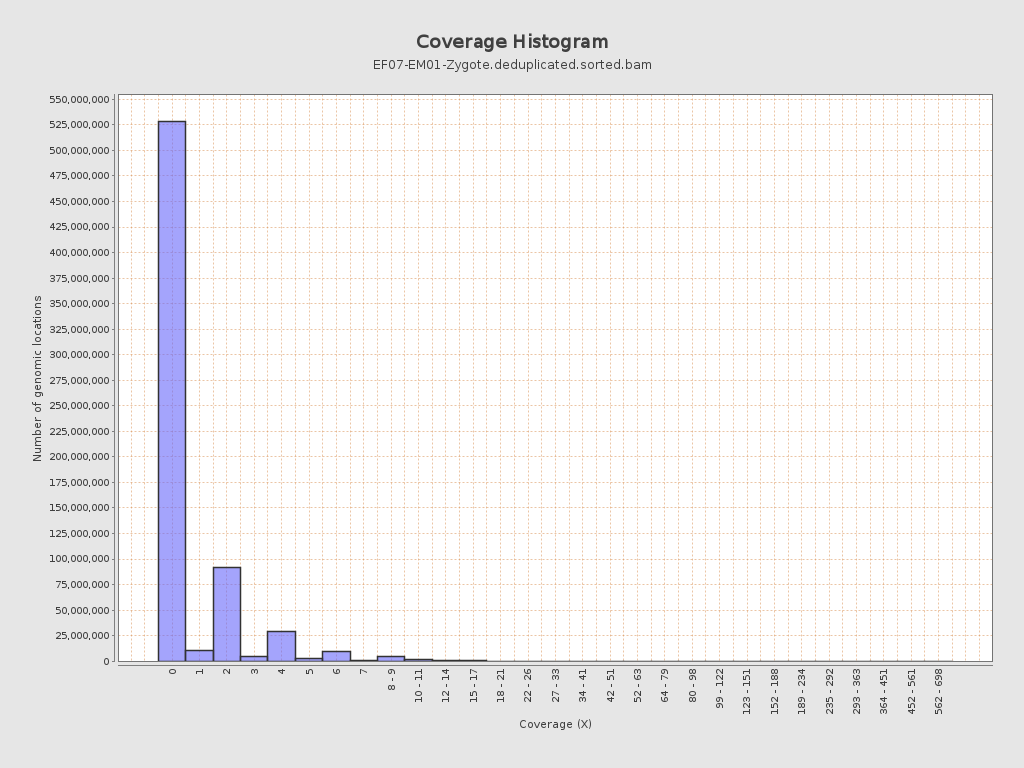
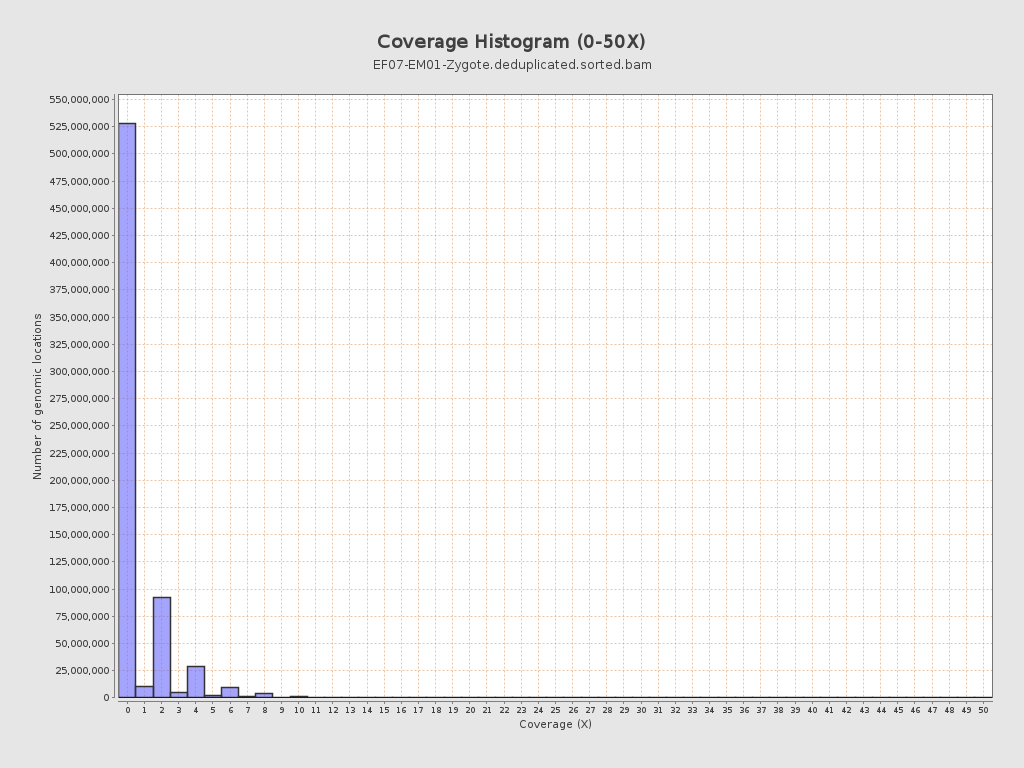
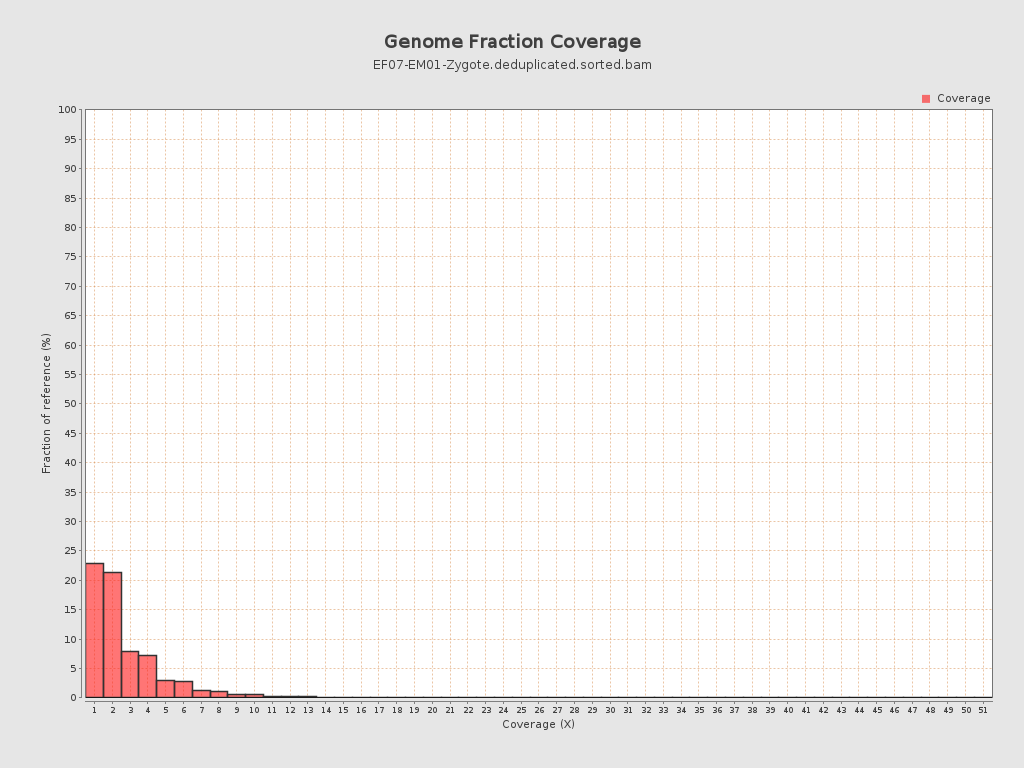
Coverage
| Mean | 0.7109 |
| Standard Deviation | 2.1349 |
| Mean (paired-end reads overlap ignored) | 0.37 |
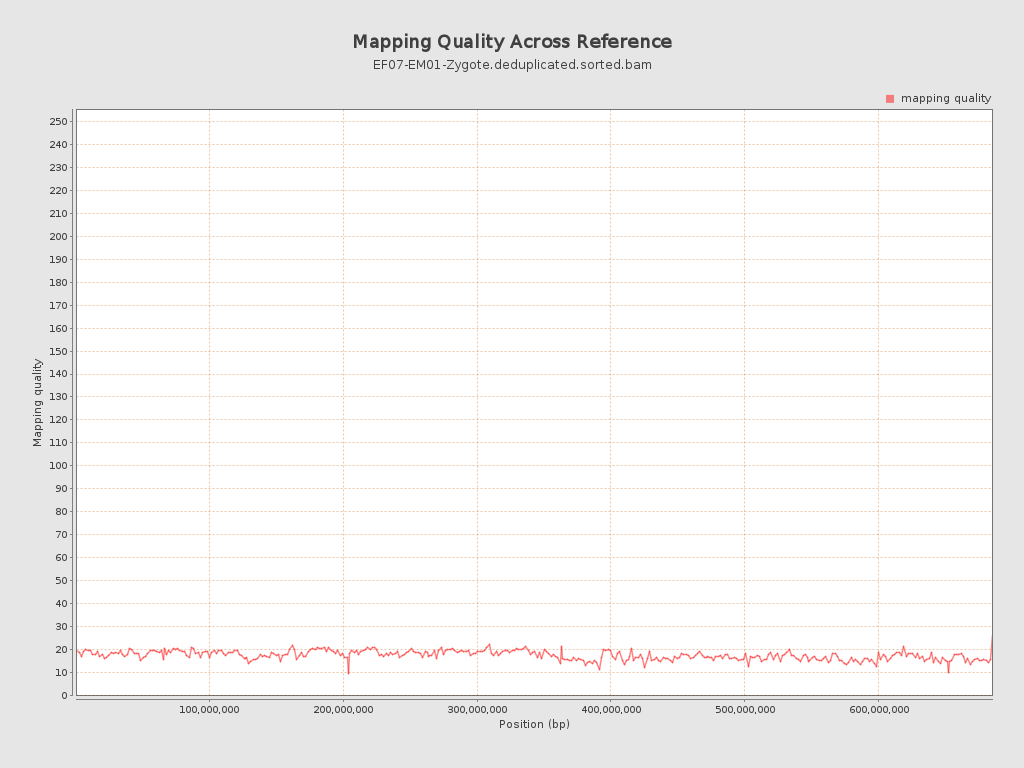
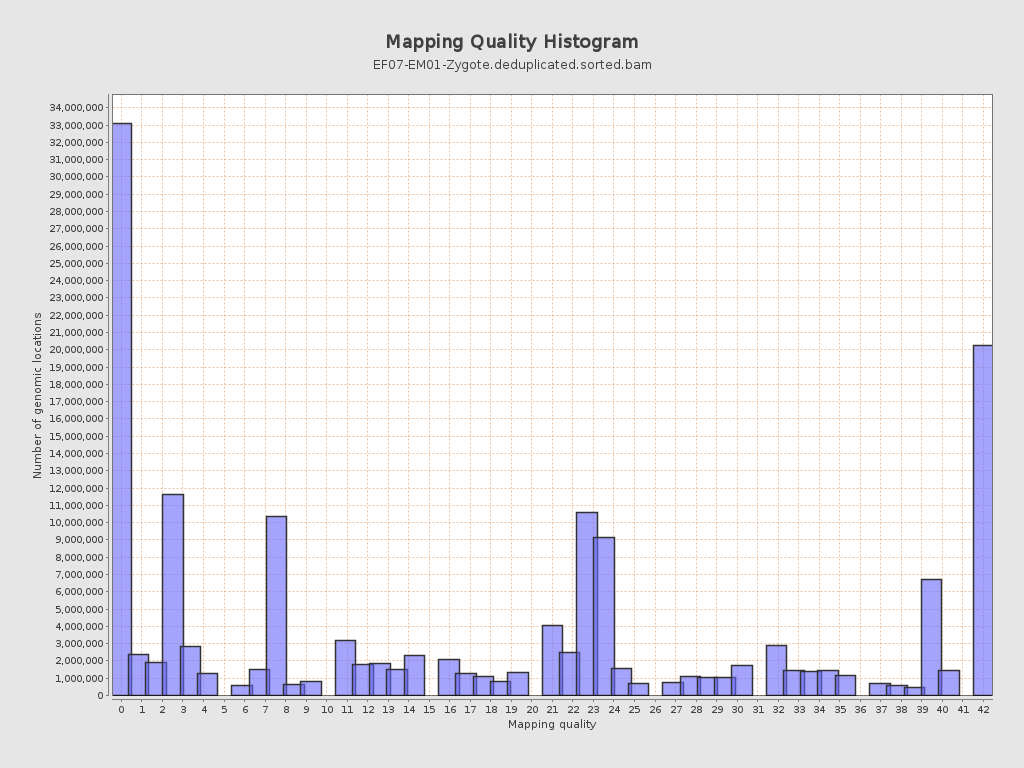
Mapping Quality
| Mean Mapping Quality | 17.5 |
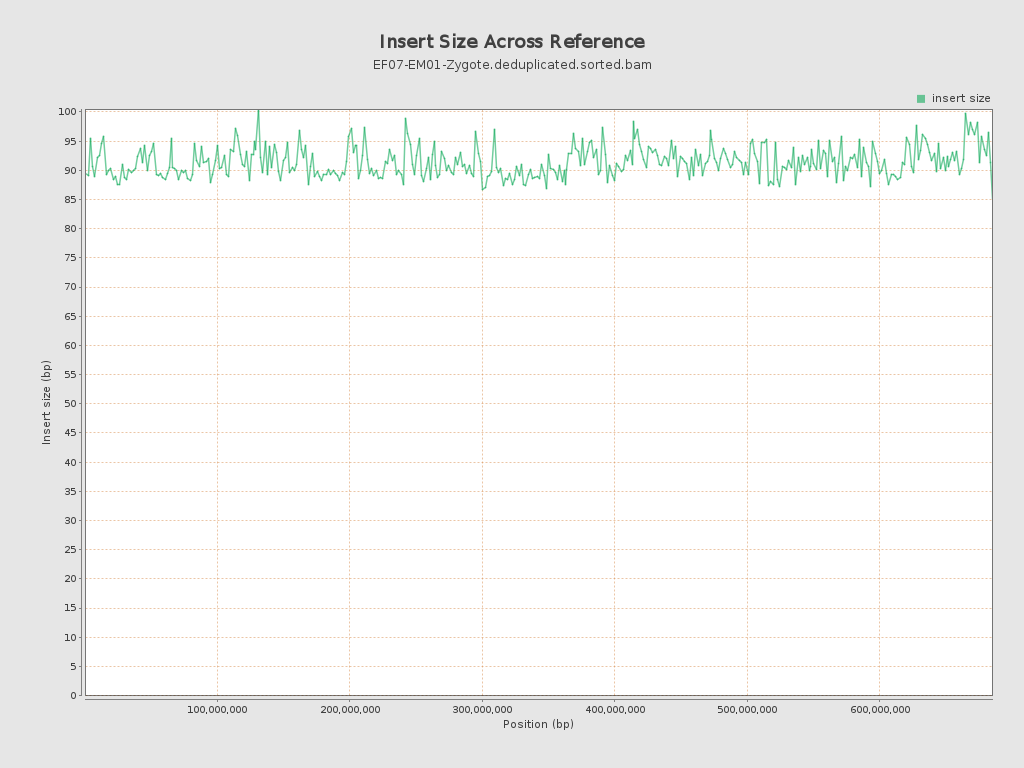
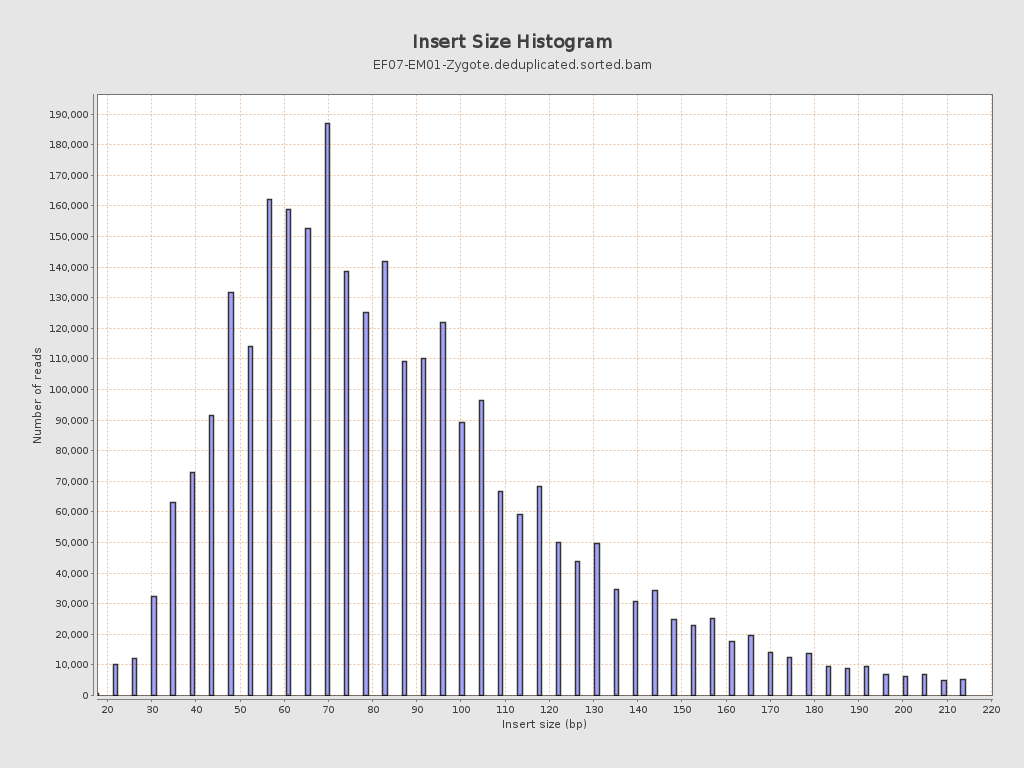
Insert size
| Mean | 90.9 |
| Standard Deviation | 43.97 |
| P25/Median/P75 | 62 / 81 / 109 |
Mismatches and indels
| General error rate | 16.91% |
| Mismatches | 81,704,073 |
| Insertions | 413,646 |
| Mapped reads with at least one insertion | 7.08% |
| Deletions | 521,521 |
| Mapped reads with at least one deletion | 8.95% |
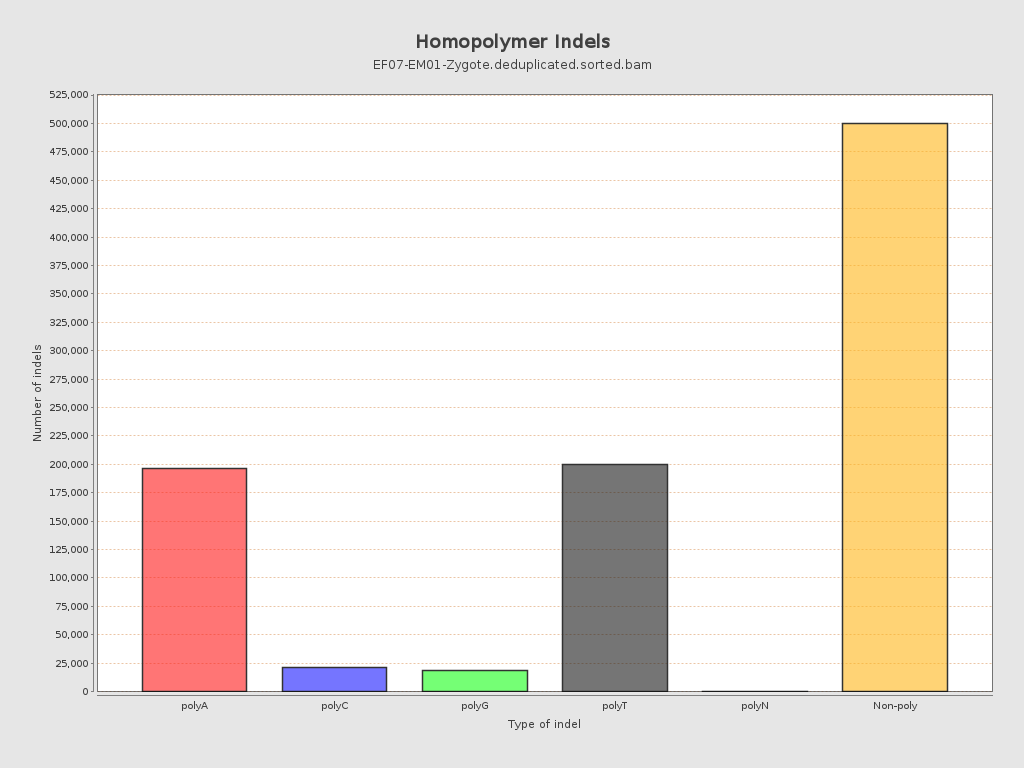
| Homopolymer indels | 46.52% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_035780.1 | 65668440 | 53818618 | 0.8196 | 1.9028 |
| NC_035781.1 | 61752955 | 55966356 | 0.9063 | 1.9842 |
| NC_035782.1 | 77061148 | 62959149 | 0.817 | 1.9468 |
| NC_035783.1 | 59691872 | 53936989 | 0.9036 | 2.0549 |
| NC_035784.1 | 98698416 | 93531573 | 0.9477 | 2.1154 |
| NC_035785.1 | 51258098 | 26110906 | 0.5094 | 1.6164 |
| NC_035786.1 | 57830854 | 29695739 | 0.5135 | 1.6808 |
| NC_035787.1 | 75944018 | 39495609 | 0.5201 | 1.4668 |
| NC_035788.1 | 104168038 | 51356275 | 0.493 | 1.521 |
| NC_035789.1 | 32650045 | 16503731 | 0.5055 | 1.594 |
| NC_007175.2 | 17244 | 3400633 | 197.2067 | 102.2579 |