Input data and parameters
QualiMap command line
| qualimap bamqc -bam EF06-EM01-Larvae.deduplicated.sorted.bam -nw 400 -hm 3 |
Alignment
| Command line: | "bismark -1 EF06-EM01-Larvae_1_val_1.fq.gz -2 EF06-EM01-Larvae_2_val_2.fq.gz --genome BismarkIndex --bam --bowtie2 --multicore 4" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | yes |
| Program: | Bismark (v0.24.2) |
| Analysis date: | Thu Dec 05 00:33:17 GMT 2024 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | EF06-EM01-Larvae.deduplicated.sorted.bam |
Summary
Globals
| Reference size | 684,741,128 |
| Number of reads | 16,759,160 |
| Mapped reads | 16,759,160 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 16,759,160 / 100% |
| Mapped reads, first in pair | 8,379,580 / 50% |
| Mapped reads, second in pair | 8,379,580 / 50% |
| Mapped reads, both in pair | 16,759,160 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Secondary alignments | 0 |
| Read min/max/mean length | 20 / 150 / 110.47 |
| Overlapping read pairs | 8,163,622 / 97.42% |
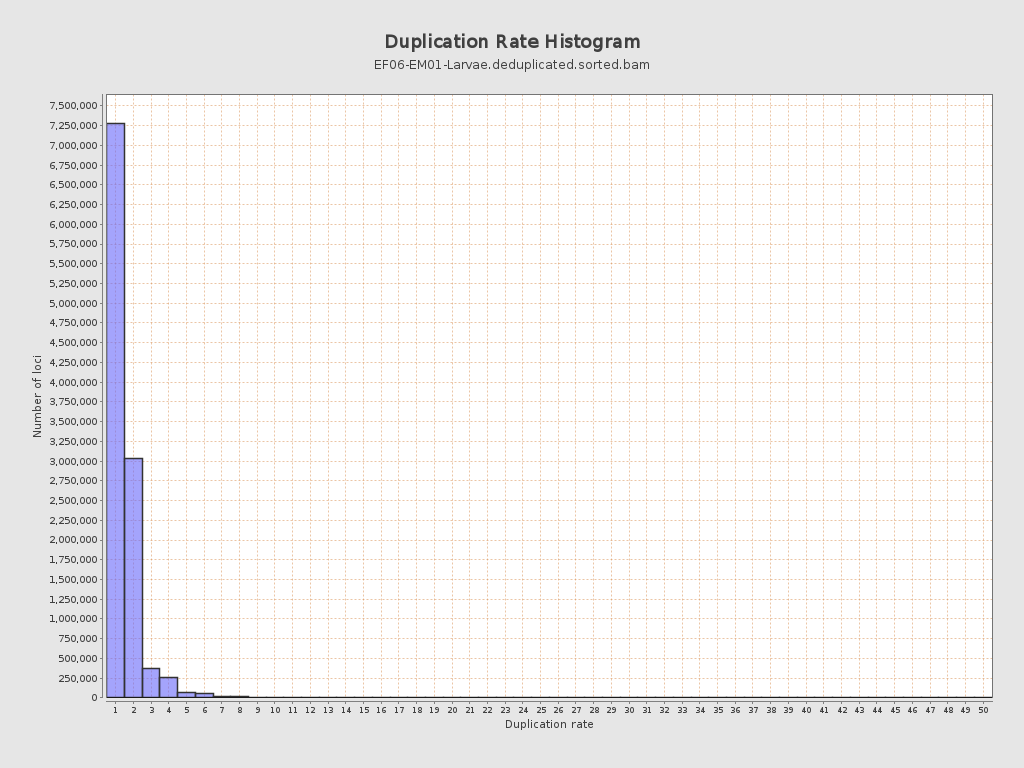
| Duplicated reads (estimated) | 5,630,883 / 33.6% |
| Duplication rate | 34.62% |
| Clipped reads | 0 / 0% |
ACGT Content
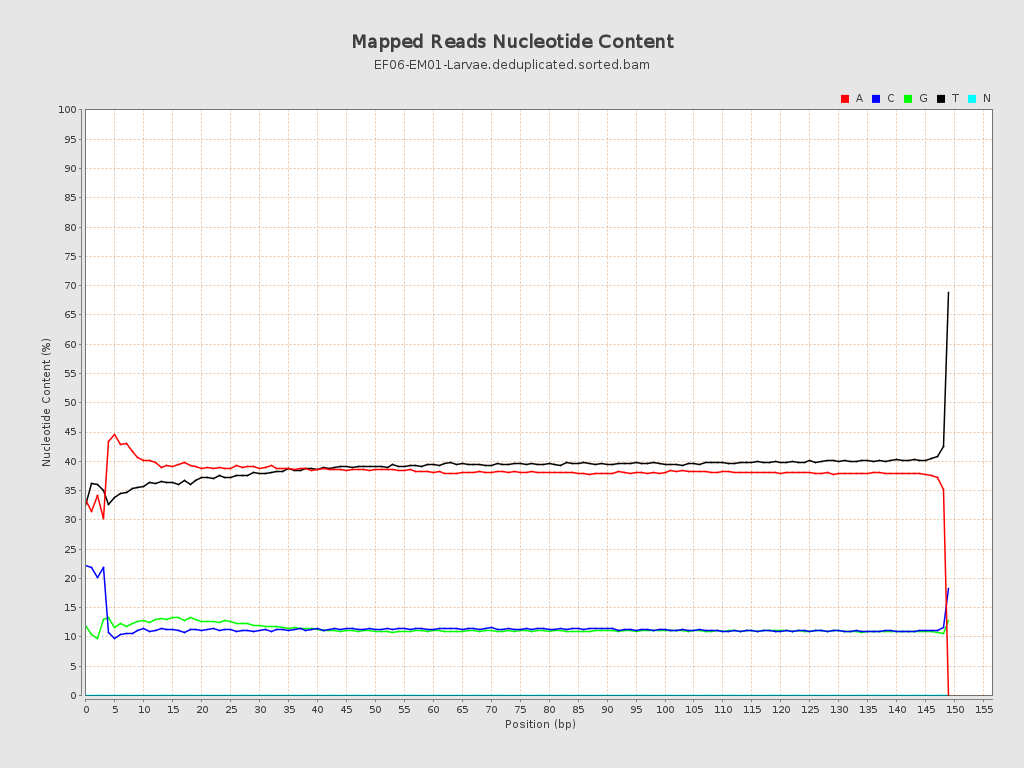
| Number/percentage of A's | 710,186,209 / 38.4% |
| Number/percentage of C's | 214,332,868 / 11.59% |
| Number/percentage of T's | 712,663,027 / 38.54% |
| Number/percentage of G's | 212,186,233 / 11.47% |
| Number/percentage of N's | 10,089 / 0% |
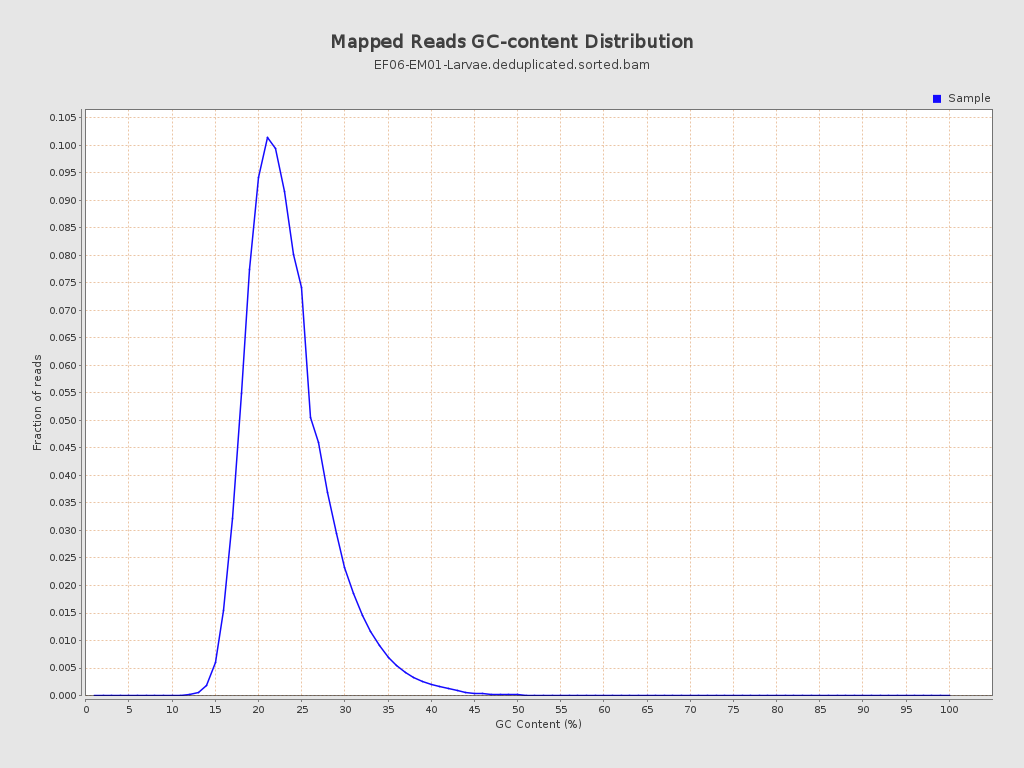
| GC Percentage | 23.06% |
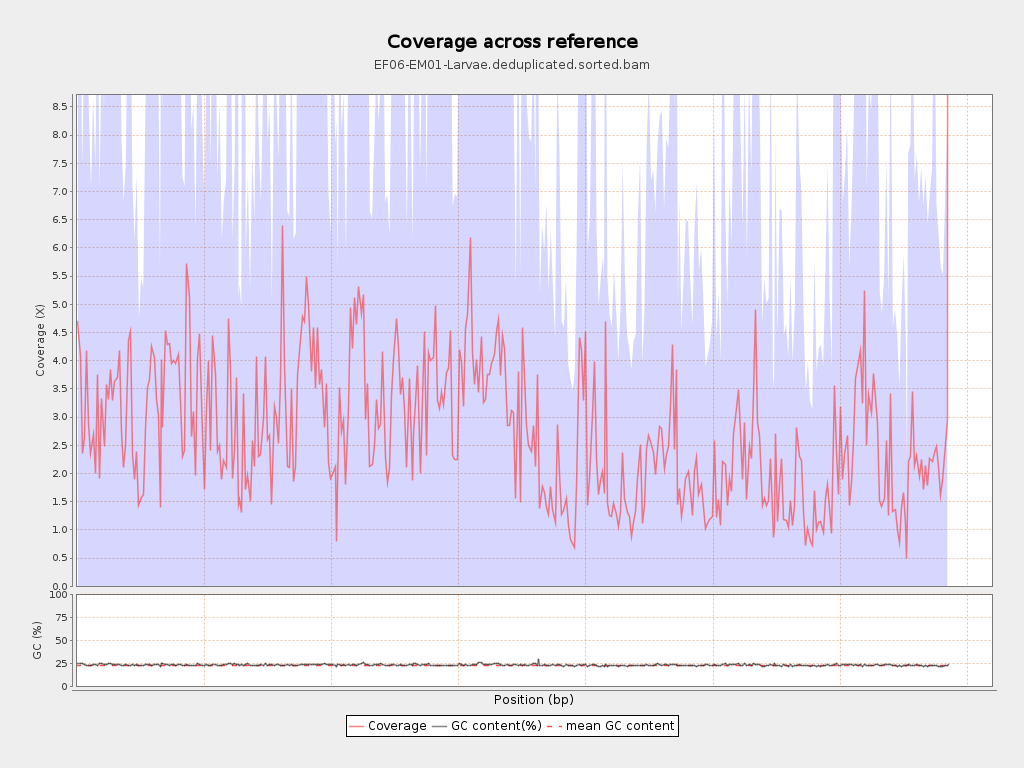
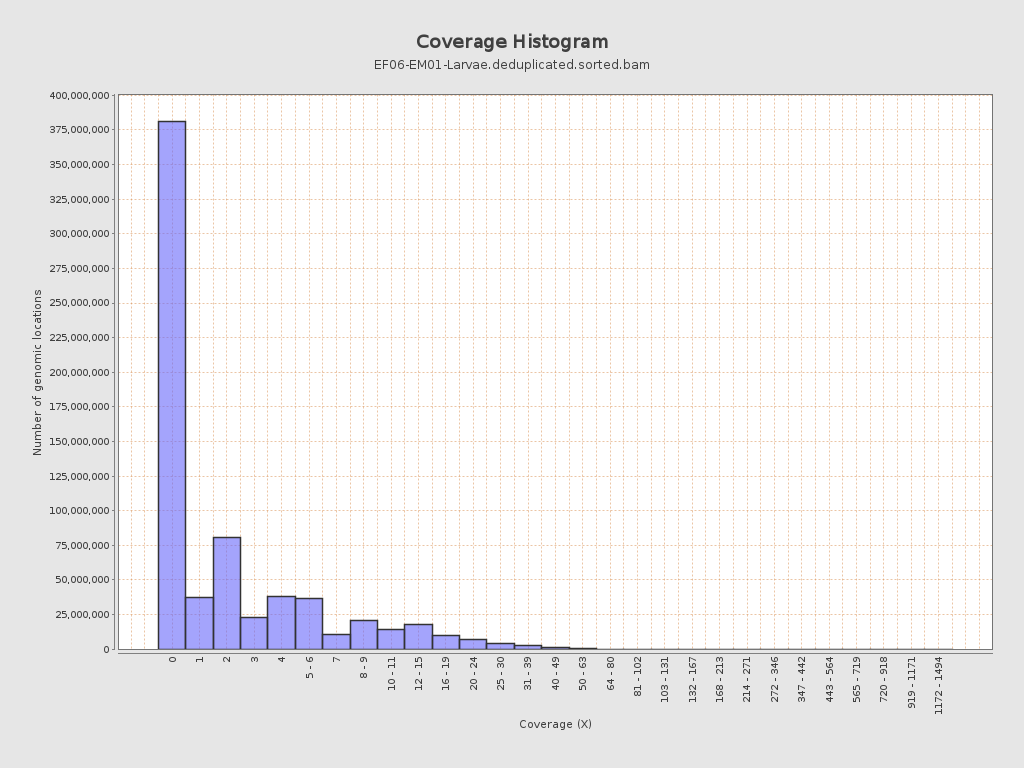
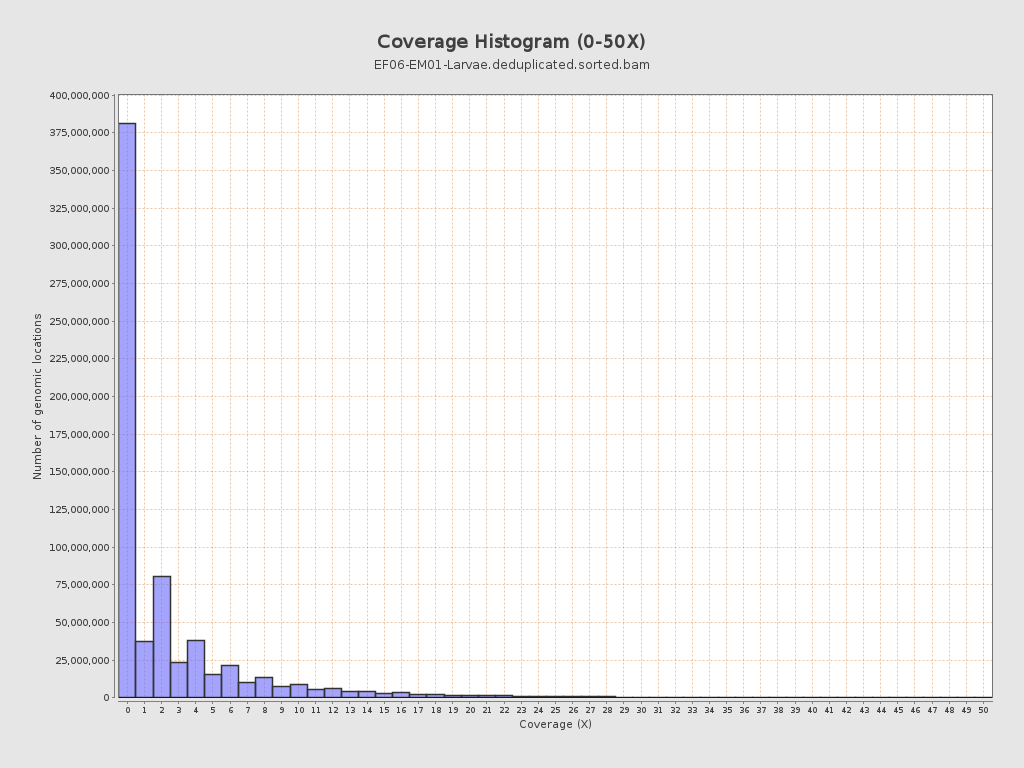
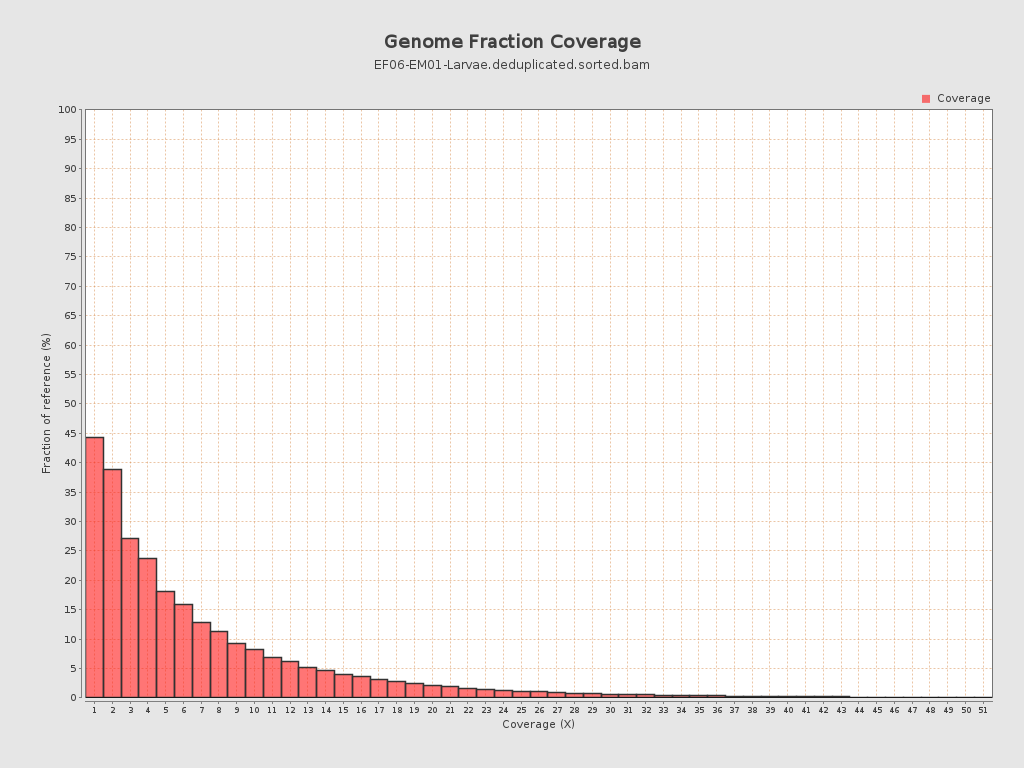
Coverage
| Mean | 2.7045 |
| Standard Deviation | 6.0894 |
| Mean (paired-end reads overlap ignored) | 1.55 |
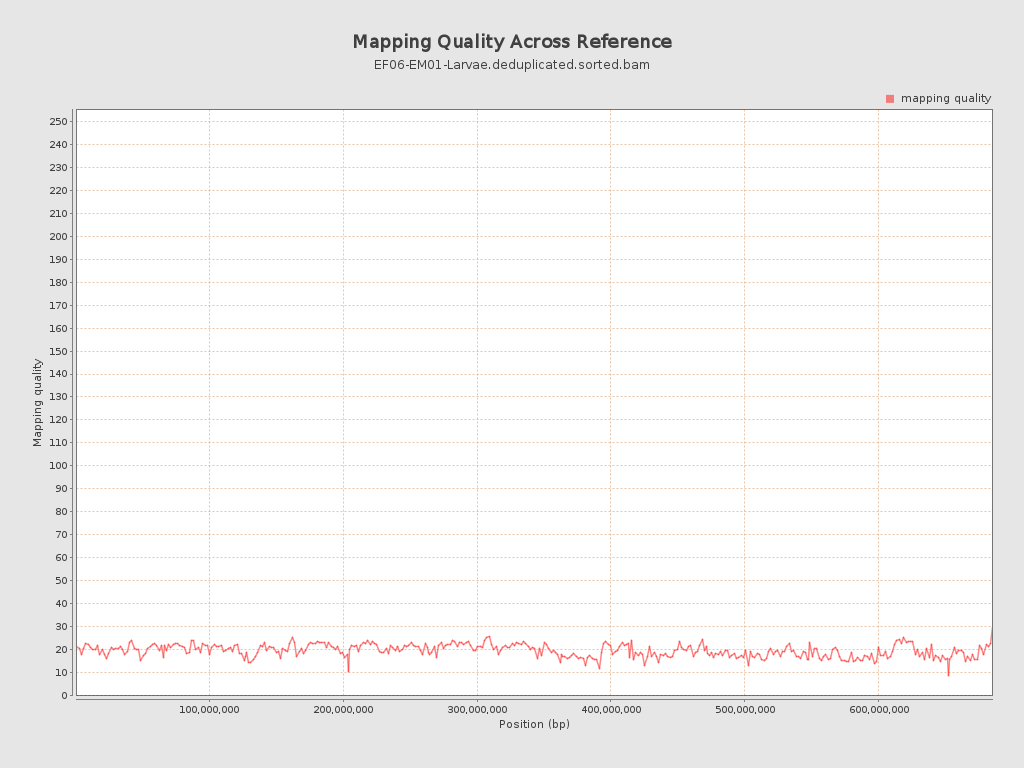
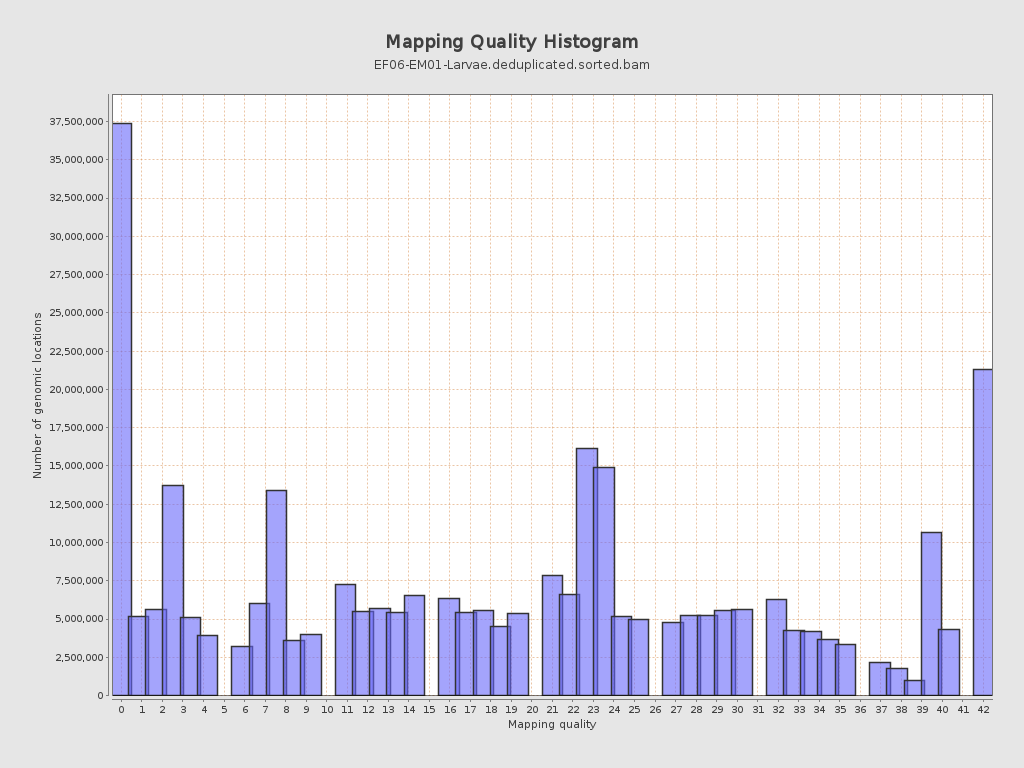
Mapping Quality
| Mean Mapping Quality | 19.54 |
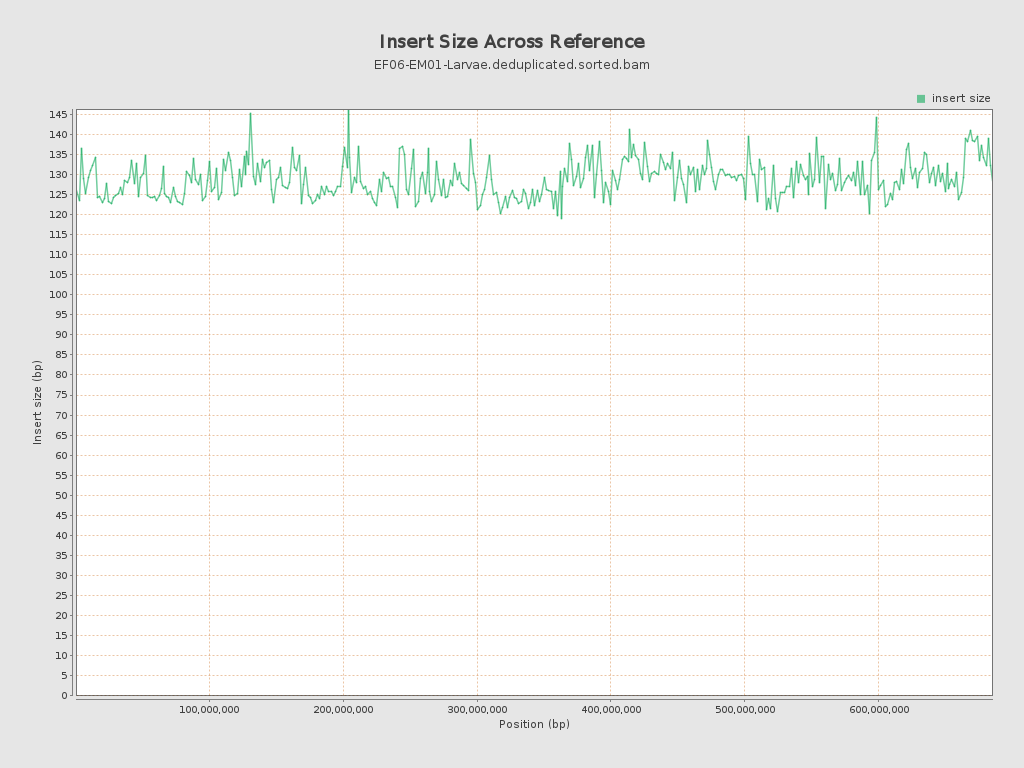
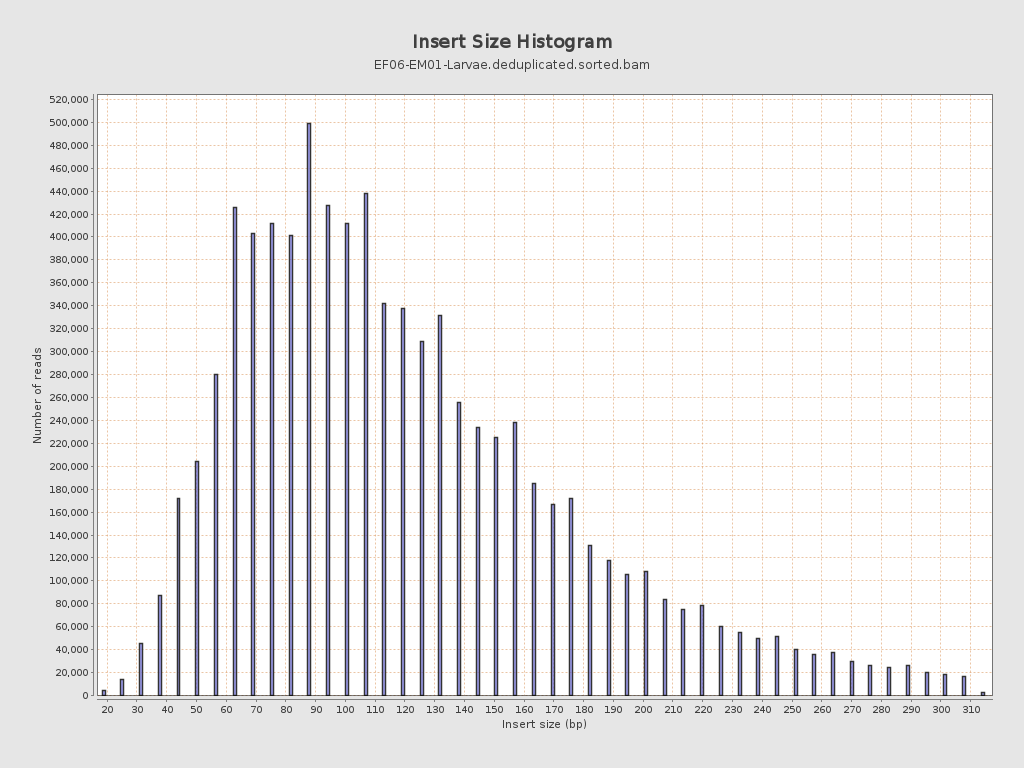
Insert size
| Mean | 128.02 |
| Standard Deviation | 65.01 |
| P25/Median/P75 | 82 / 113 / 157 |
Mismatches and indels
| General error rate | 19.28% |
| Mismatches | 354,910,716 |
| Insertions | 1,357,995 |
| Mapped reads with at least one insertion | 7.67% |
| Deletions | 1,838,987 |
| Mapped reads with at least one deletion | 10.48% |
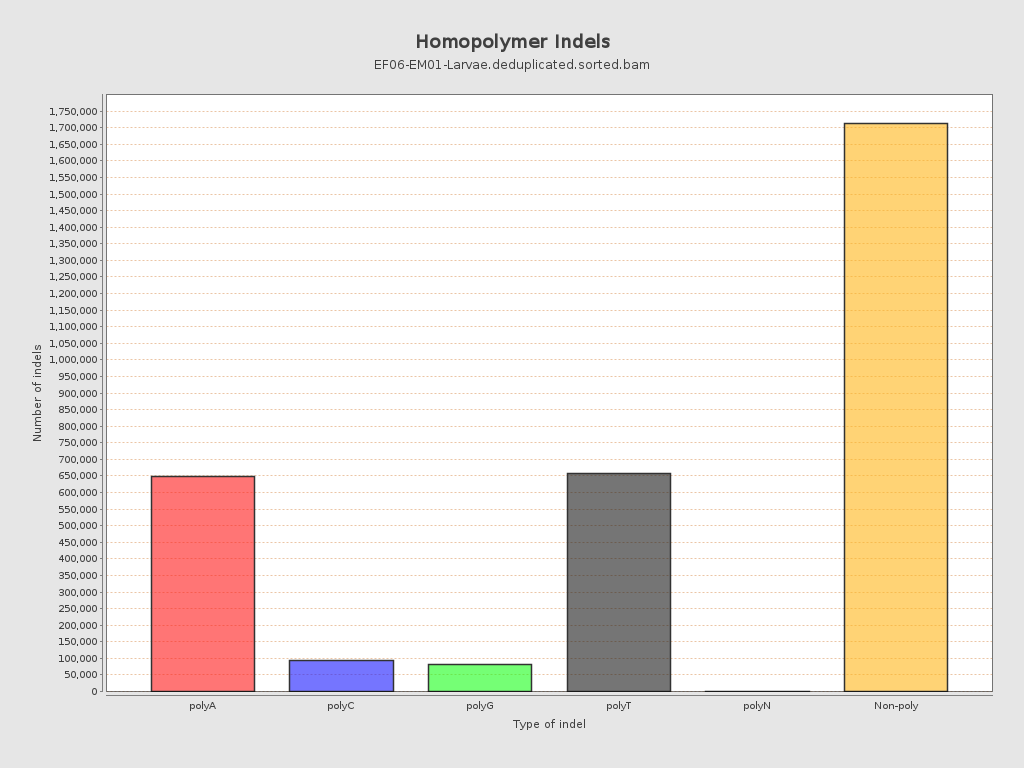
| Homopolymer indels | 46.41% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_035780.1 | 65668440 | 201051366 | 3.0616 | 6.2282 |
| NC_035781.1 | 61752955 | 205757054 | 3.3319 | 7.0503 |
| NC_035782.1 | 77061148 | 238418213 | 3.0939 | 6.835 |
| NC_035783.1 | 59691872 | 201252434 | 3.3715 | 6.3434 |
| NC_035784.1 | 98698416 | 349364604 | 3.5397 | 7.1647 |
| NC_035785.1 | 51258098 | 103261503 | 2.0145 | 4.7647 |
| NC_035786.1 | 57830854 | 120224246 | 2.0789 | 4.6587 |
| NC_035787.1 | 75944018 | 149473656 | 1.9682 | 4.8975 |
| NC_035788.1 | 104168038 | 208147211 | 1.9982 | 5.4342 |
| NC_035789.1 | 32650045 | 73003276 | 2.2359 | 5.0865 |
| NC_007175.2 | 17244 | 1928018 | 111.808 | 55.3886 |