Input data and parameters
QualiMap command line
| qualimap bamqc -bam CF05-CM05-Zygote.deduplicated.sorted.bam -nw 400 -hm 3 |
Alignment
| Command line: | "bismark -1 CF05-CM05-Zygote_1_val_1.fq.gz -2 CF05-CM05-Zygote_2_val_2.fq.gz --genome BismarkIndex --bam --bowtie2 --multicore 4" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | yes |
| Program: | Bismark (v0.24.2) |
| Analysis date: | Thu Dec 05 00:30:54 GMT 2024 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | CF05-CM05-Zygote.deduplicated.sorted.bam |
Summary
Globals
| Reference size | 684,741,128 |
| Number of reads | 11,511,062 |
| Mapped reads | 11,511,062 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 11,511,062 / 100% |
| Mapped reads, first in pair | 5,755,531 / 50% |
| Mapped reads, second in pair | 5,755,531 / 50% |
| Mapped reads, both in pair | 11,511,062 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Secondary alignments | 0 |
| Read min/max/mean length | 20 / 150 / 98.14 |
| Overlapping read pairs | 5,735,807 / 99.66% |
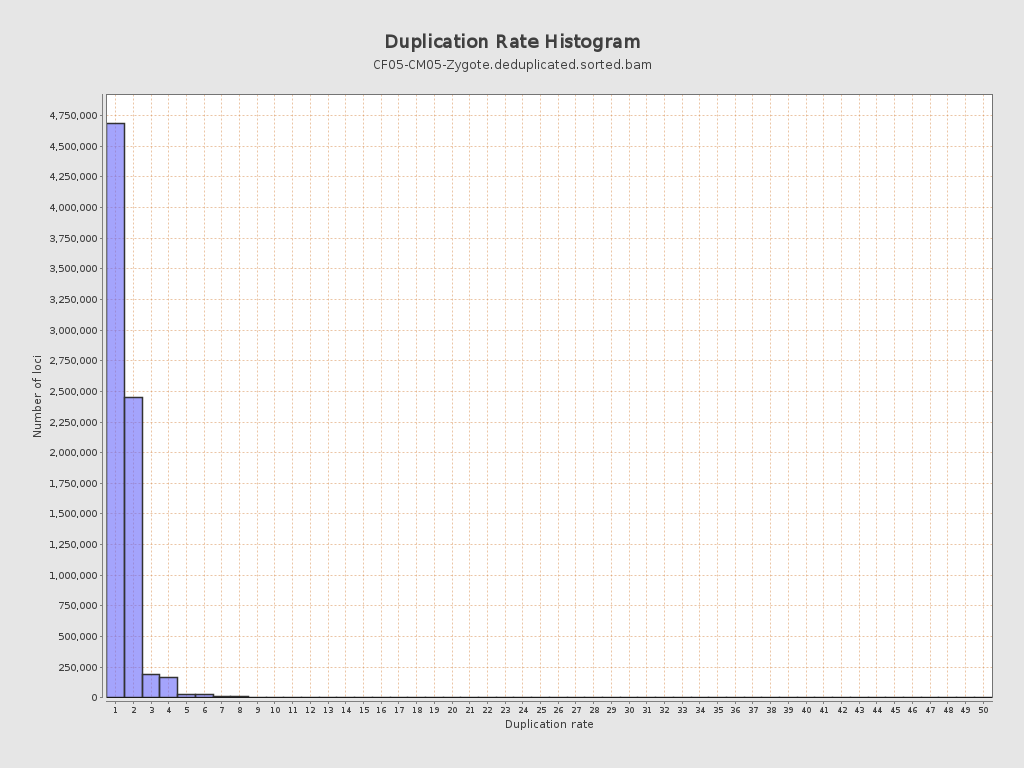
| Duplicated reads (estimated) | 3,924,615 / 34.09% |
| Duplication rate | 38.25% |
| Clipped reads | 0 / 0% |
ACGT Content
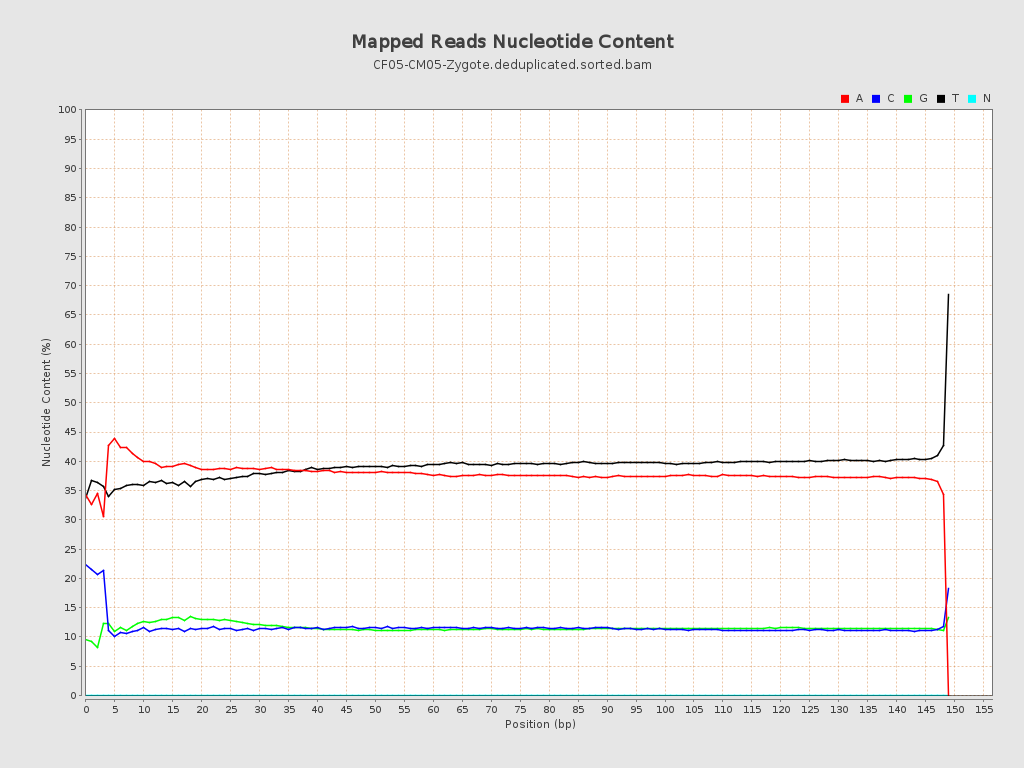
| Number/percentage of A's | 430,006,737 / 38.11% |
| Number/percentage of C's | 133,138,924 / 11.8% |
| Number/percentage of T's | 433,751,287 / 38.44% |
| Number/percentage of G's | 131,533,746 / 11.66% |
| Number/percentage of N's | 6,898 / 0% |
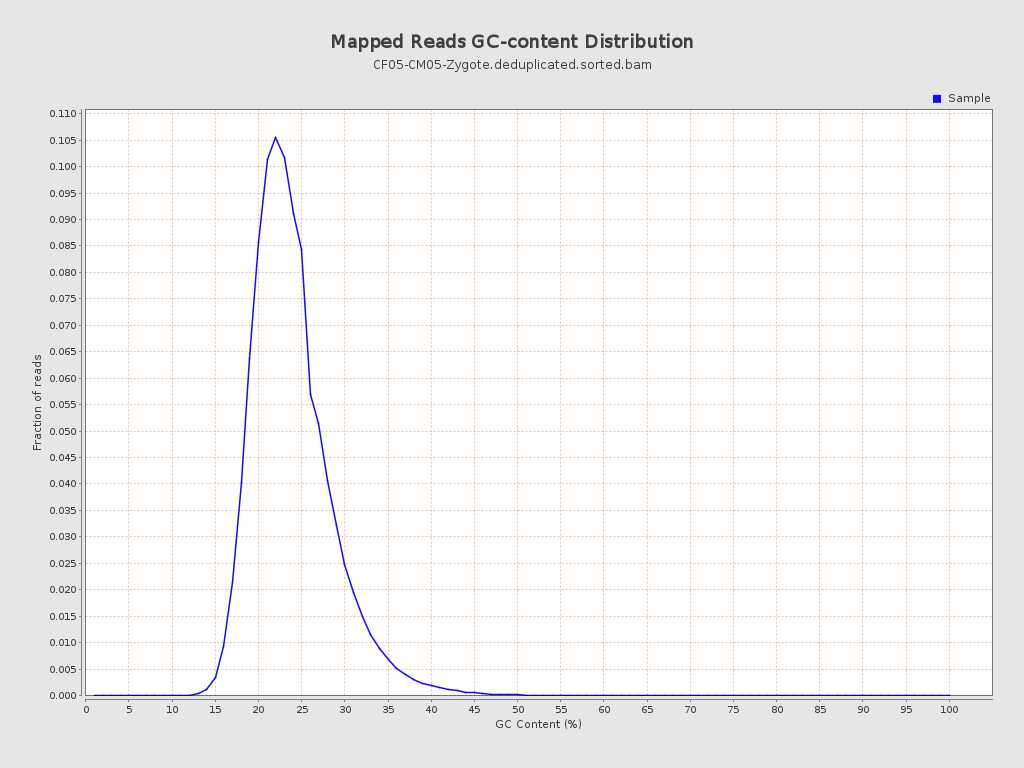
| GC Percentage | 23.45% |
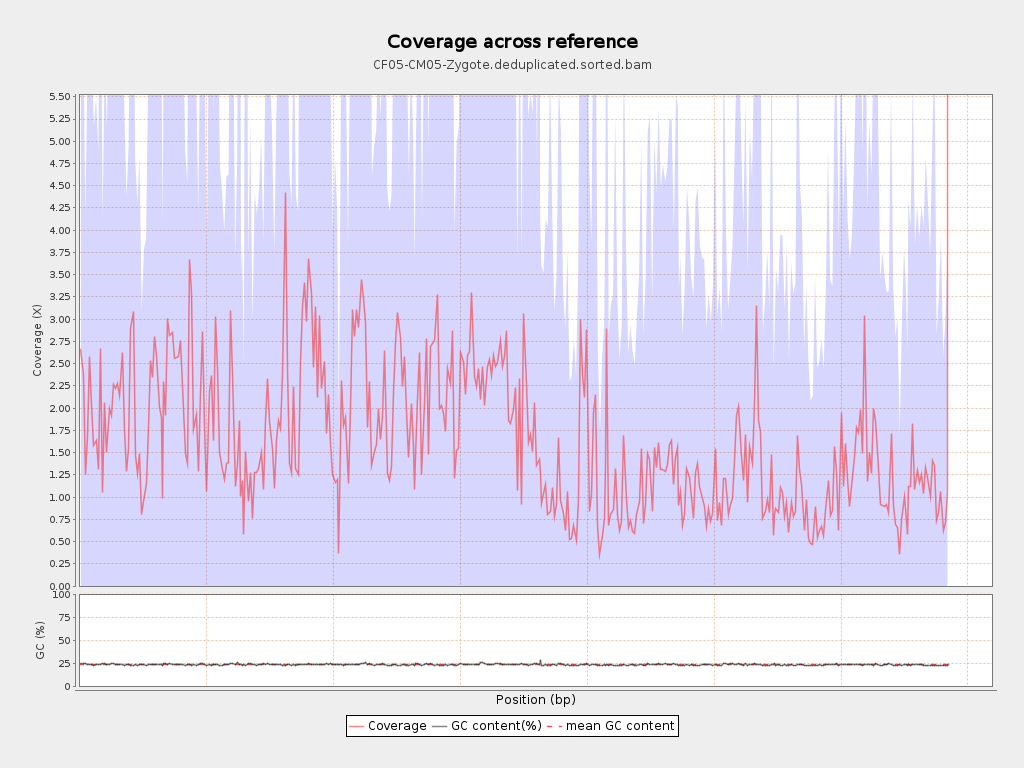
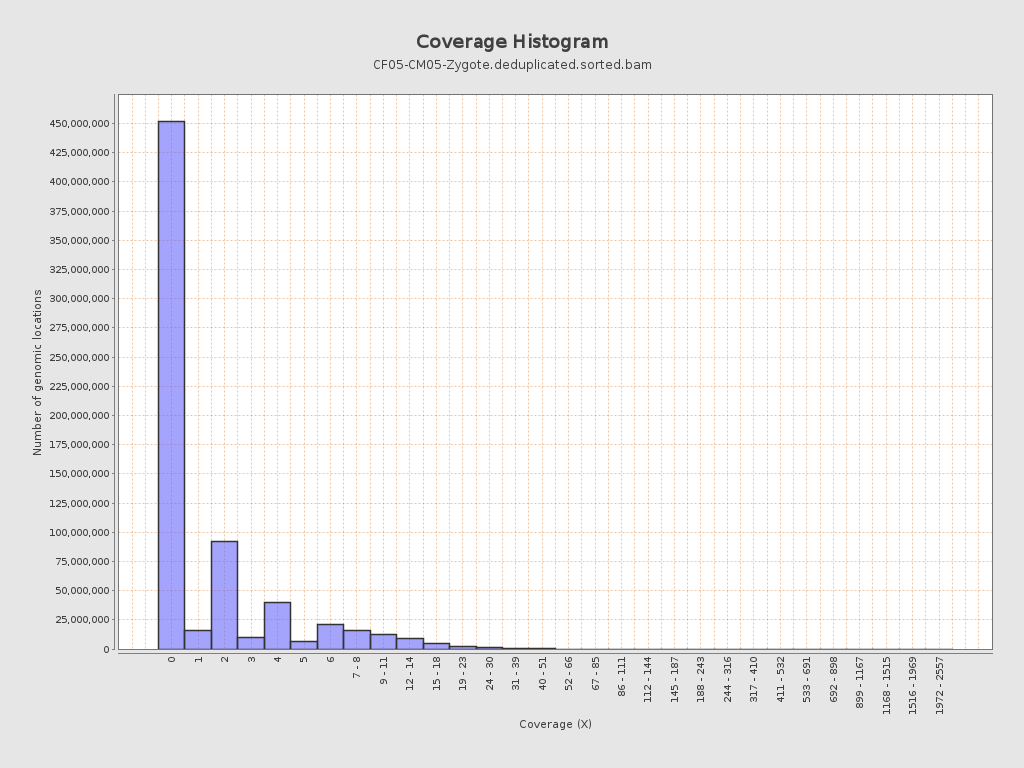
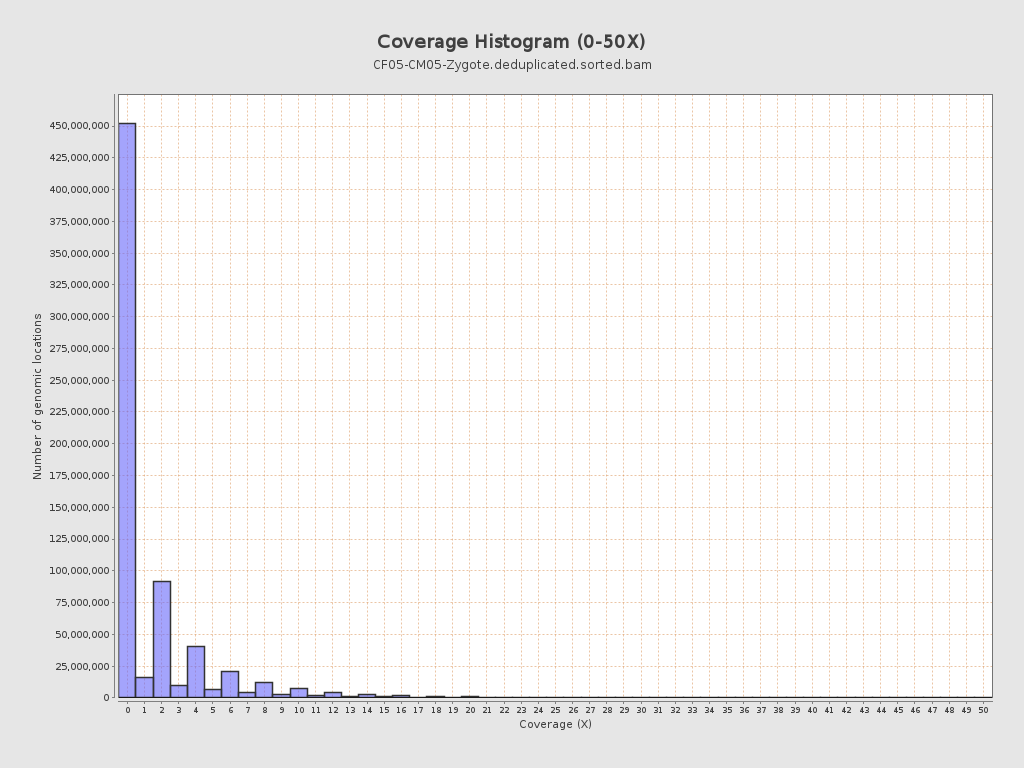
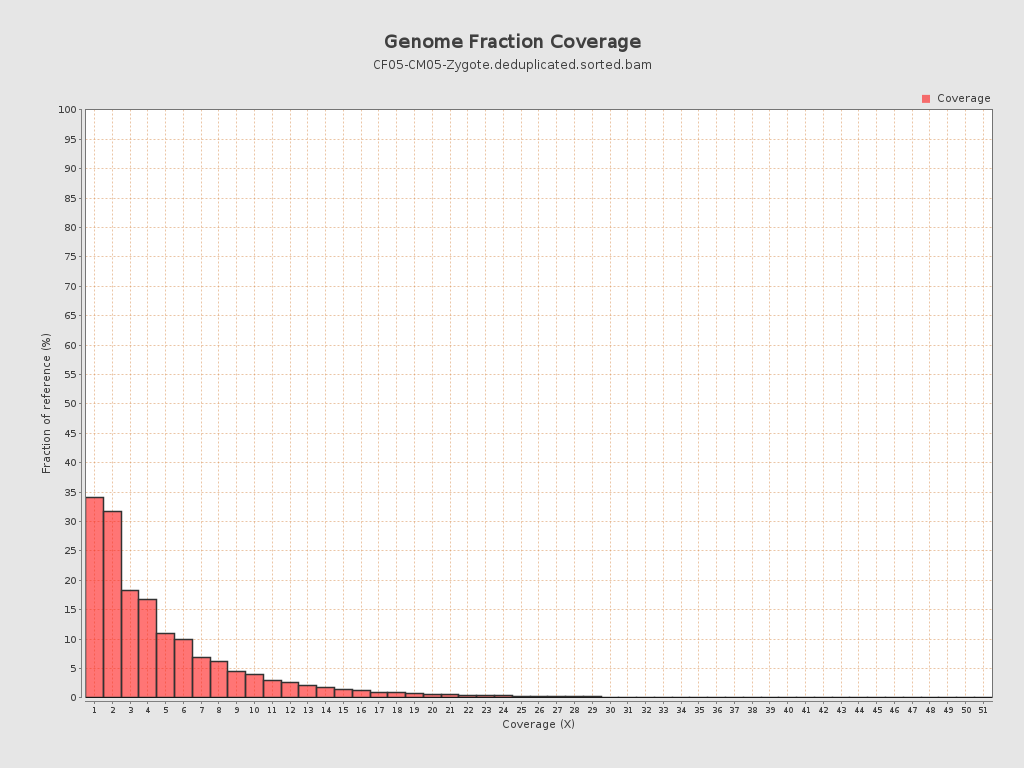
Coverage
| Mean | 1.651 |
| Standard Deviation | 5.6351 |
| Mean (paired-end reads overlap ignored) | 0.88 |
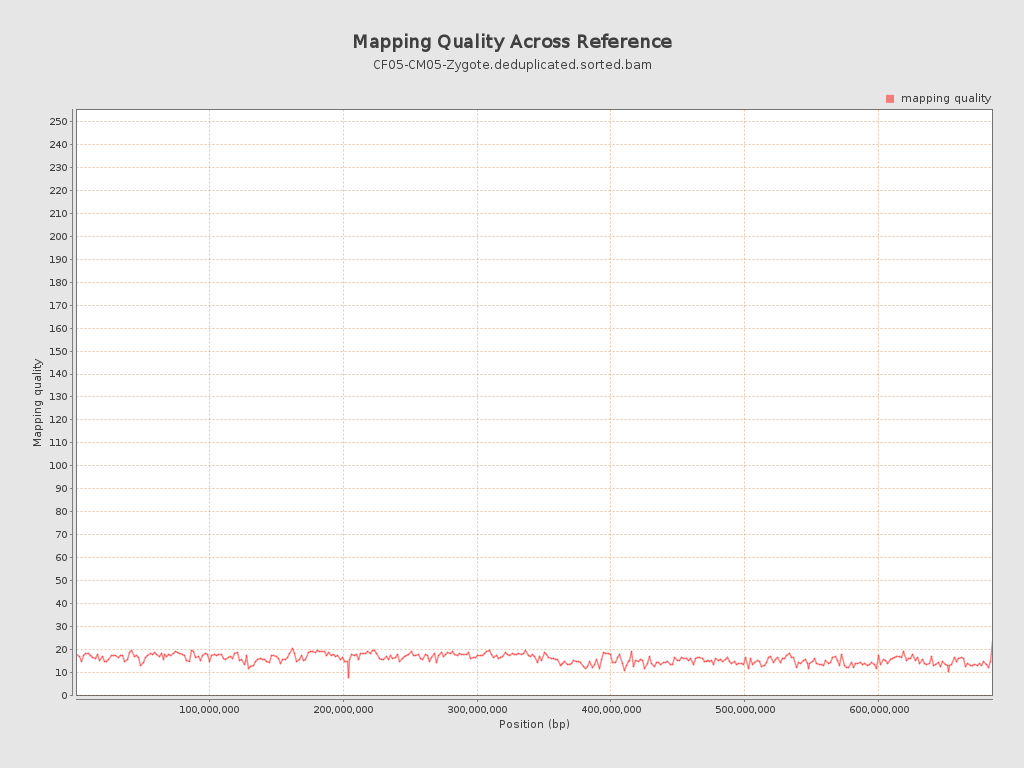
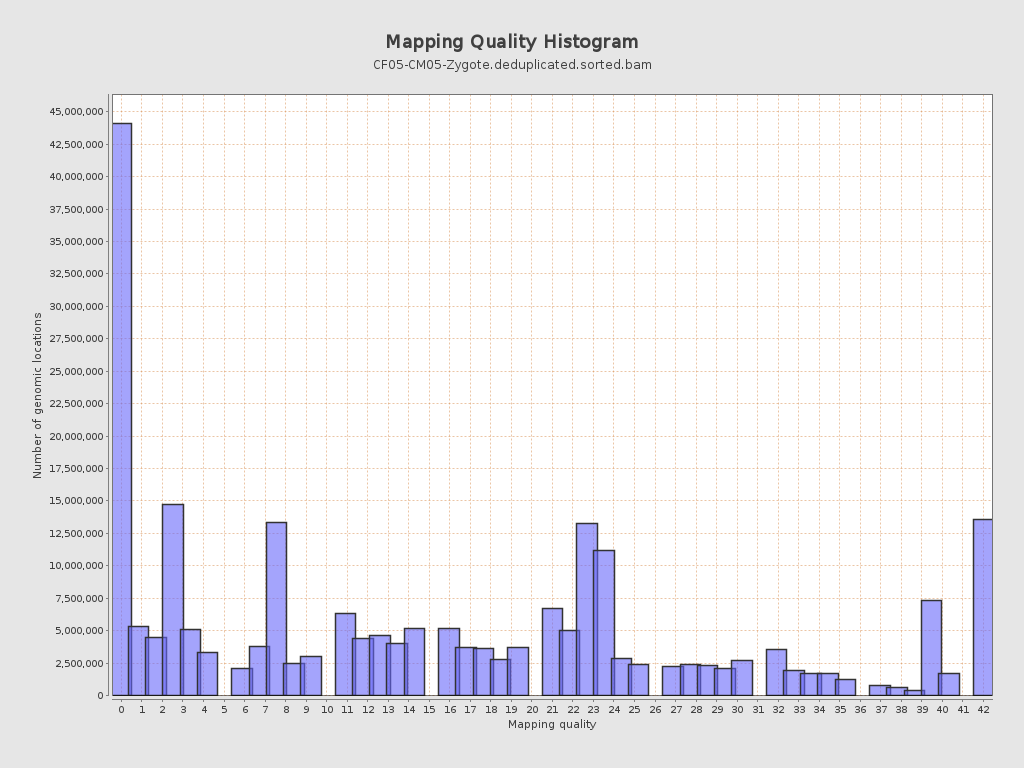
Mapping Quality
| Mean Mapping Quality | 15.96 |
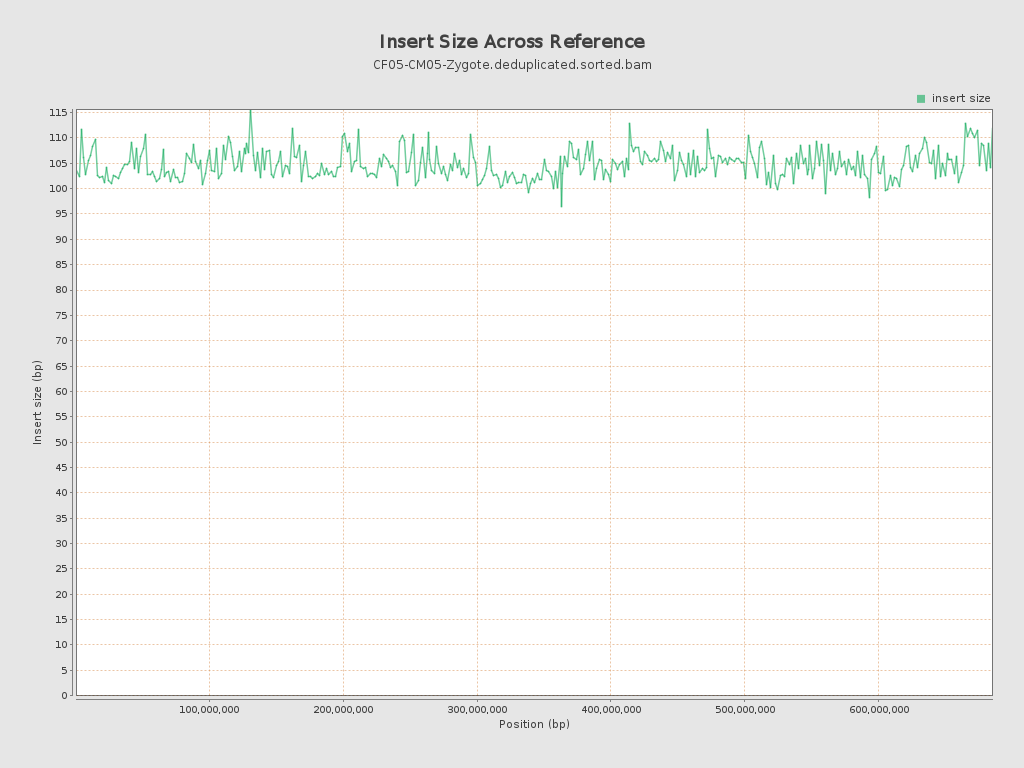
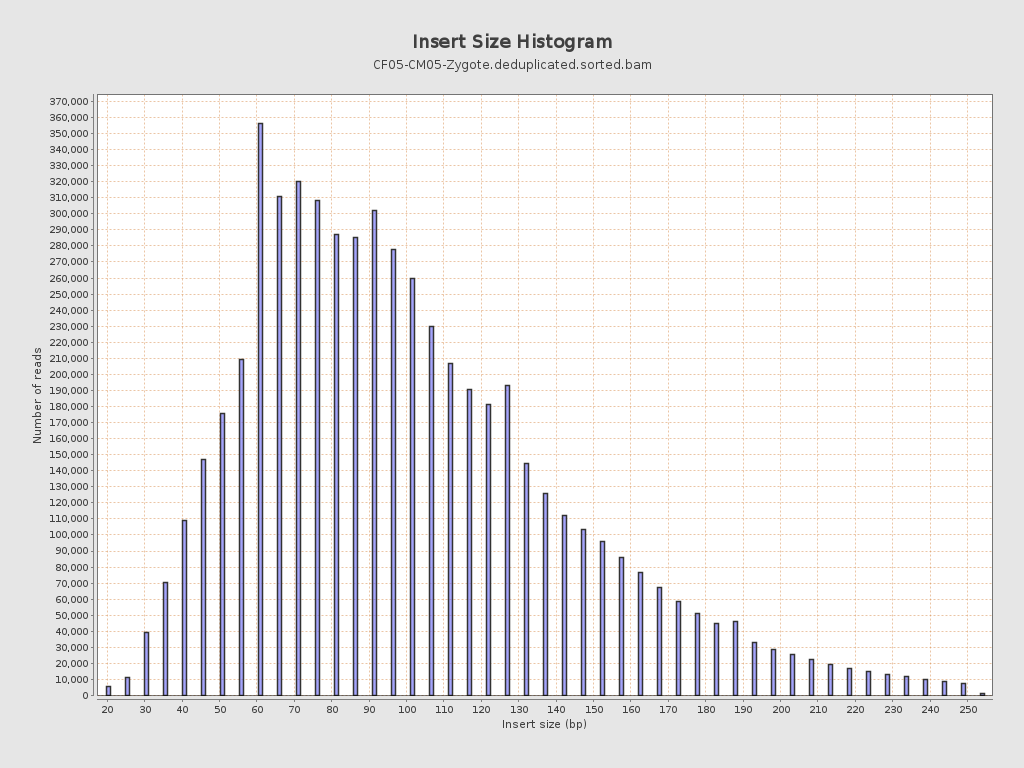
Insert size
| Mean | 104.41 |
| Standard Deviation | 45.94 |
| P25/Median/P75 | 72 / 95 / 127 |
Mismatches and indels
| General error rate | 17.98% |
| Mismatches | 202,005,750 |
| Insertions | 841,995 |
| Mapped reads with at least one insertion | 7.04% |
| Deletions | 1,574,585 |
| Mapped reads with at least one deletion | 12.95% |
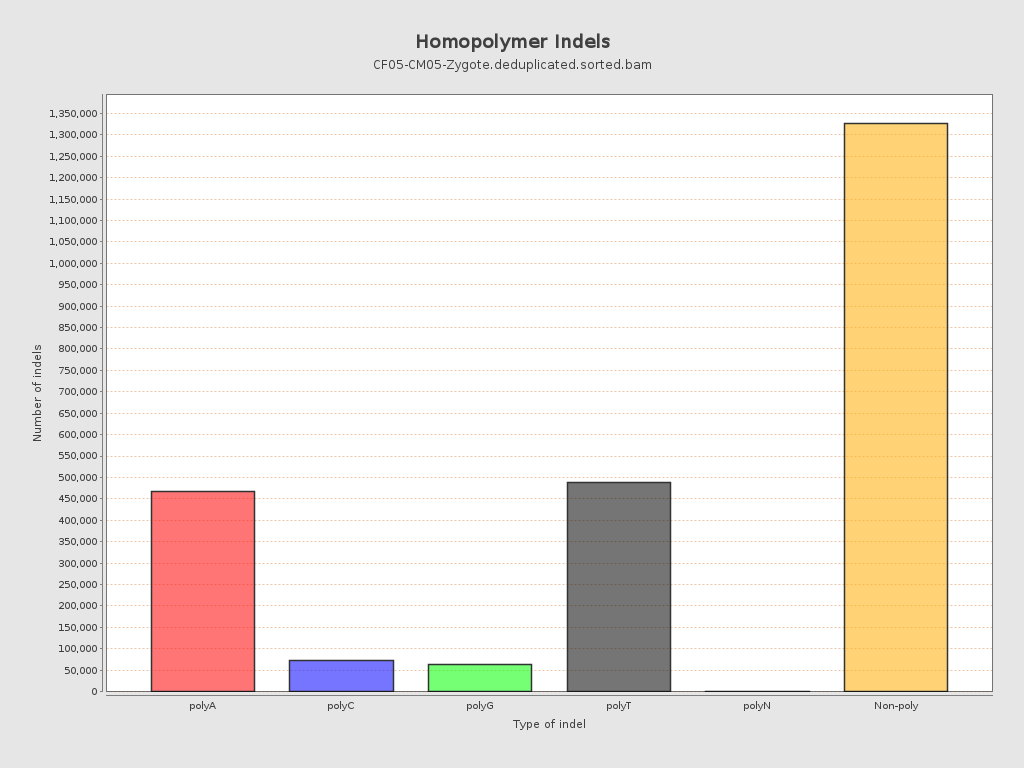
| Homopolymer indels | 45.09% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_035780.1 | 65668440 | 126509538 | 1.9265 | 3.923 |
| NC_035781.1 | 61752955 | 130496038 | 2.1132 | 4.3727 |
| NC_035782.1 | 77061148 | 148409384 | 1.9259 | 4.1069 |
| NC_035783.1 | 59691872 | 128029031 | 2.1448 | 4.2584 |
| NC_035784.1 | 98698416 | 216811195 | 2.1967 | 4.2954 |
| NC_035785.1 | 51258098 | 59267903 | 1.1563 | 3.1972 |
| NC_035786.1 | 57830854 | 67610354 | 1.1691 | 2.984 |
| NC_035787.1 | 75944018 | 90423255 | 1.1907 | 3.0242 |
| NC_035788.1 | 104168038 | 113533632 | 1.0899 | 2.9635 |
| NC_035789.1 | 32650045 | 36464089 | 1.1168 | 2.9454 |
| NC_007175.2 | 17244 | 12972556 | 752.2939 | 387.4227 |