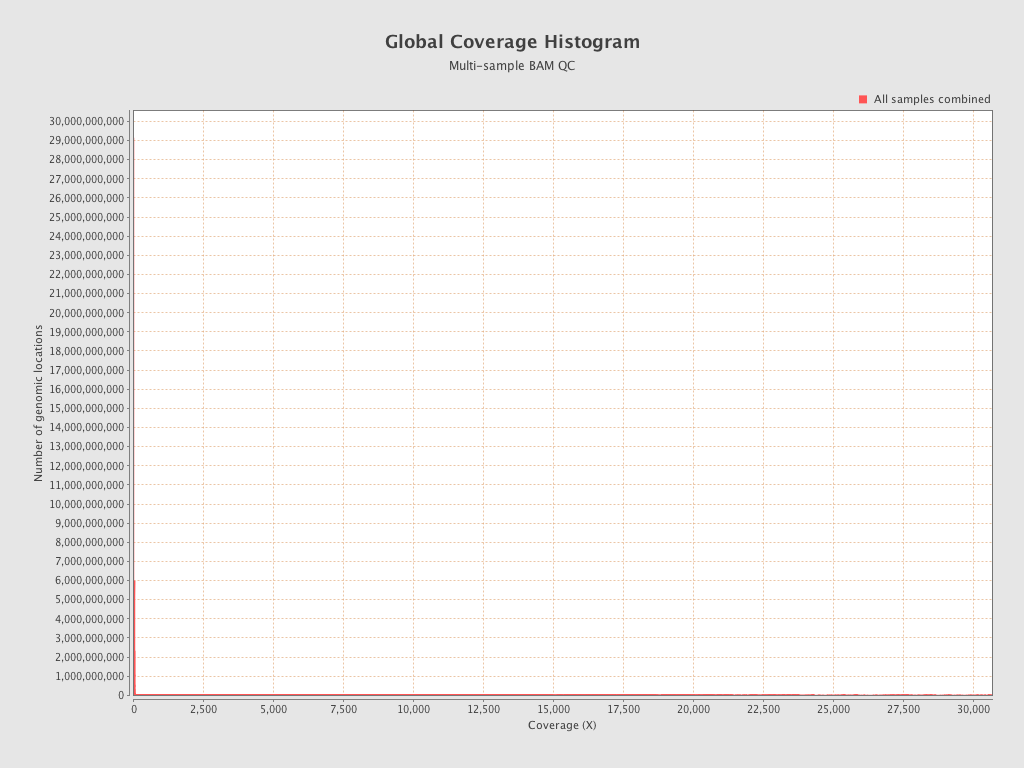
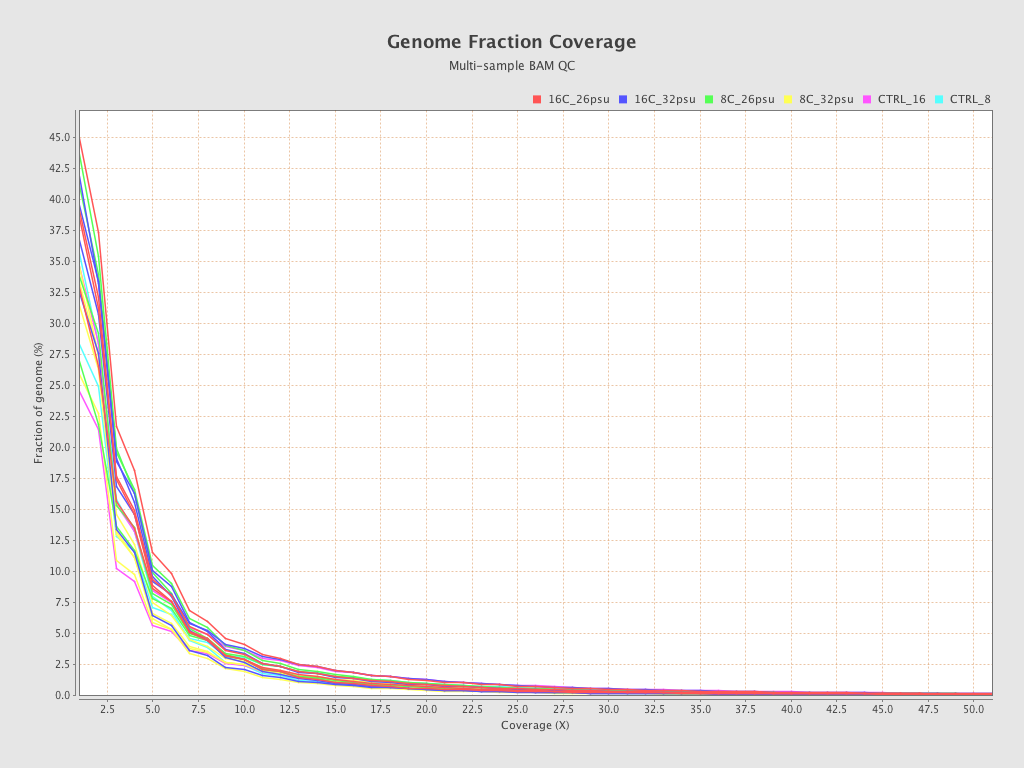
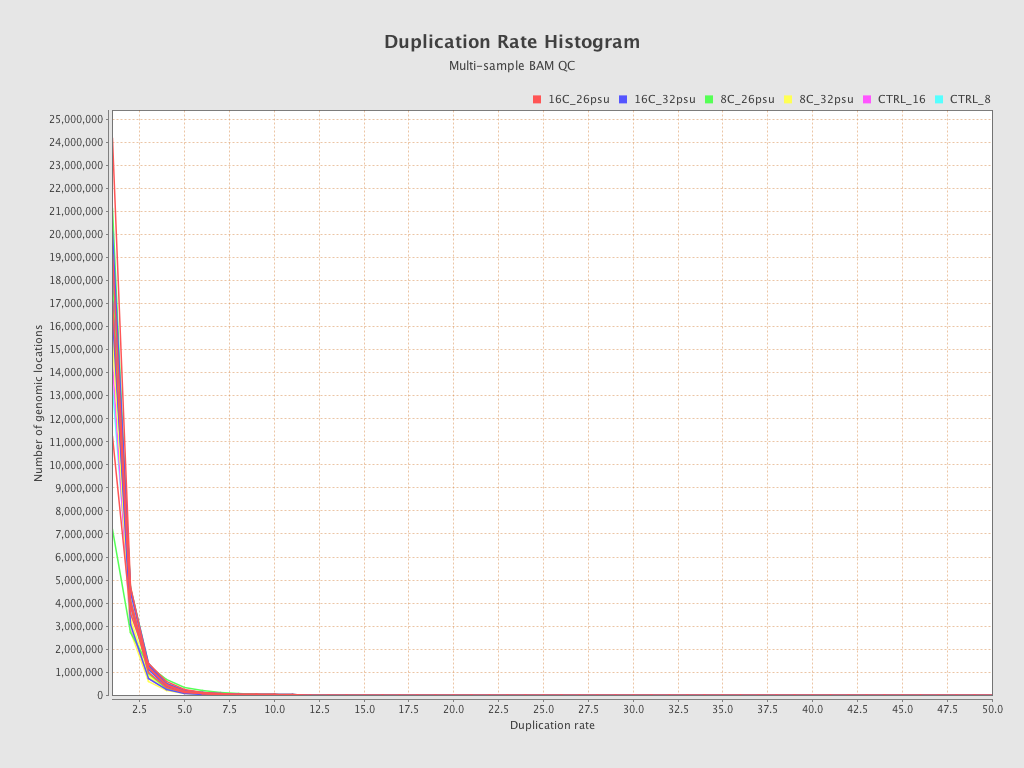
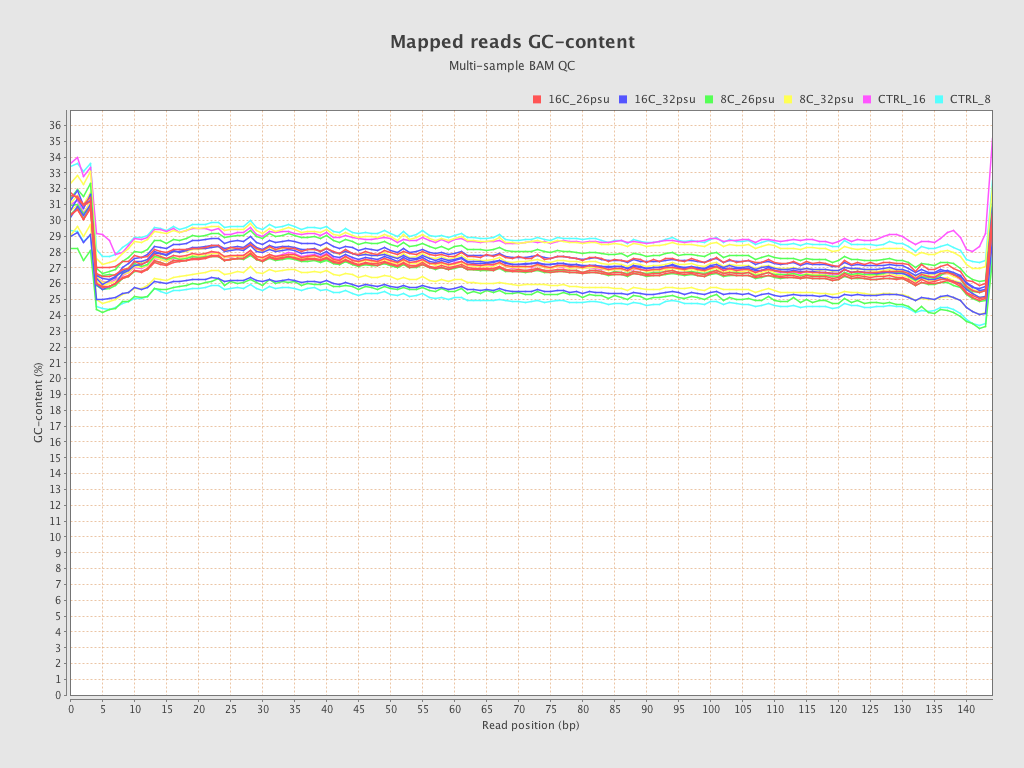
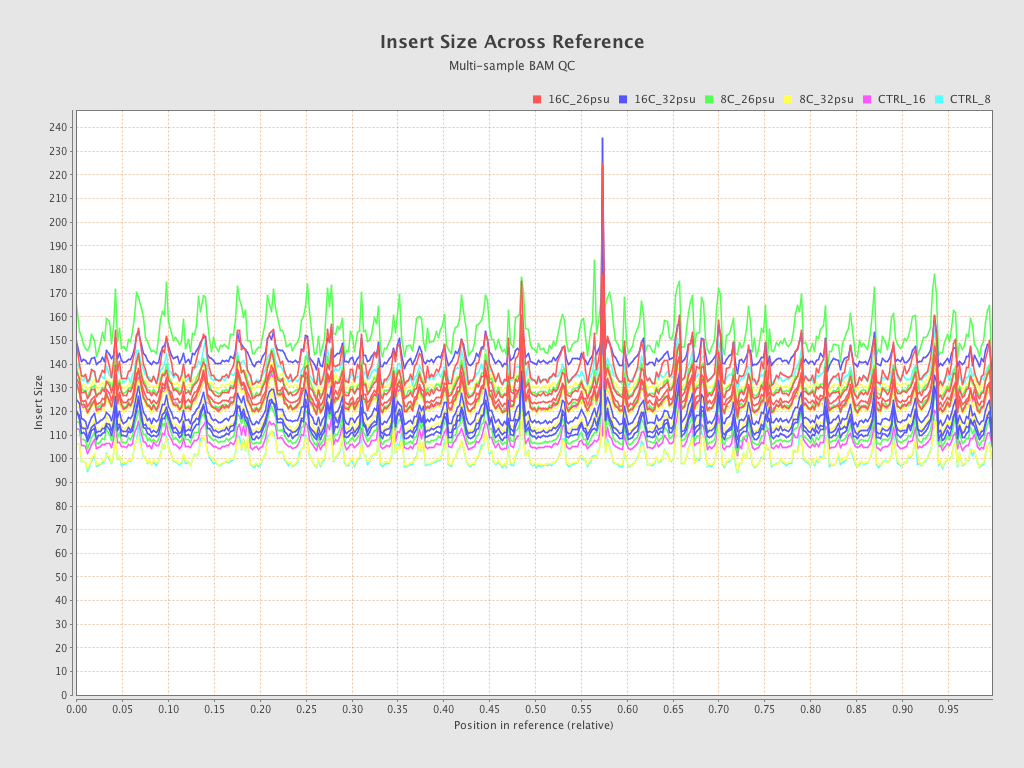
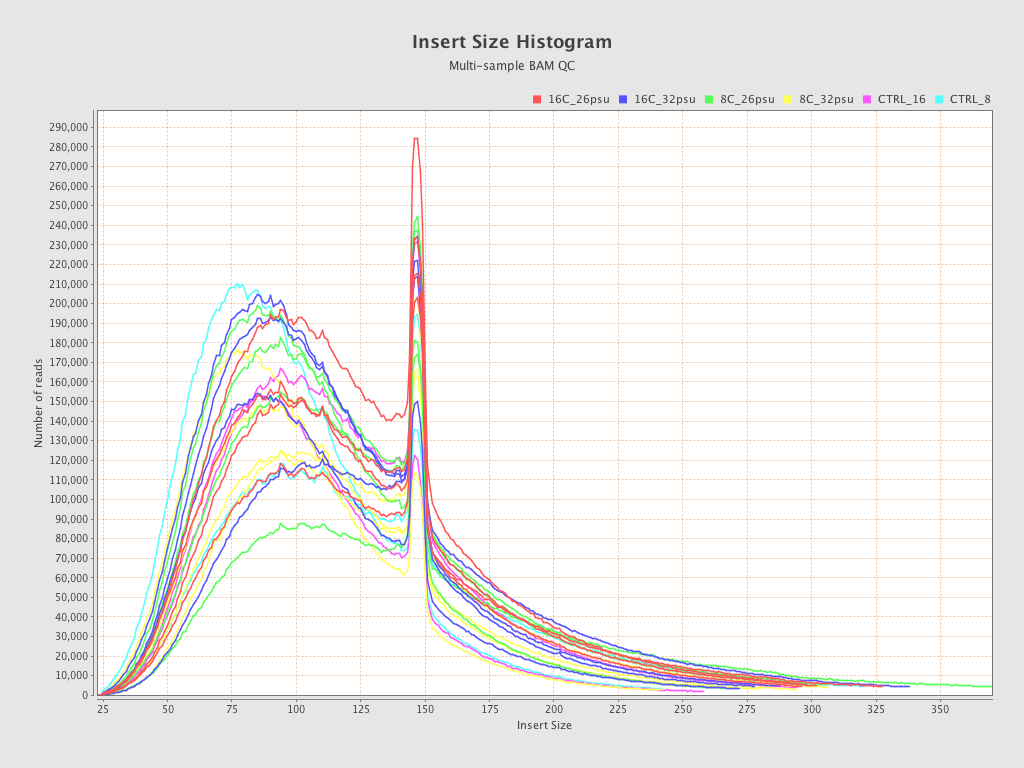
| Sample8 (16C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_32psu_4_S4.sorted_stats |
| Sample9 (8C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_26psu_1_S9.sorted_stats |
| Sample13 (8C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_32psu_1_S5.sorted_stats |
| Sample17 (CTRL_16) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/CTRL_16C_26psu_1_S19.sorted_stats |
| Sample20 (CTRL_8) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/CTRL_8C_26psu_2_S18.sorted_stats |
| Sample7 (16C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_32psu_3_S3.sorted_stats |
| Sample18 (CTRL_16) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/CTRL_16C_26psu_2_S21.sorted_stats |
| Sample4 (16C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_26psu_4_S16.sorted_stats |
| Sample11 (8C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_26psu_3_S11.sorted_stats |
| Sample3 (16C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_26psu_3_S15.sorted_stats |
| Sample6 (16C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_32psu_2_S2.sorted_stats |
| Sample15 (8C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_32psu_3_S7.sorted_stats |
| Sample19 (CTRL_8) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/CTRL_8C_26psu_1_S17.sorted_stats |
| Sample5 (16C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_32psu_1_S1.sorted_stats |
| Sample1 (16C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_26psu_1_S13.sorted_stats |
| Sample2 (16C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/16C_26psu_2_S14.sorted_stats |
| Sample14 (8C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_32psu_2_S6.sorted_stats |
| Sample16 (8C_32psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_32psu_4_S8.sorted_stats |
| Sample10 (8C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_26psu_2_S10.sorted_stats |
| Sample12 (8C_26psu) |
/Volumes/web/metacarcinus/Salmo_Calig/analyses/20190828_Salmo/8C_26psu_4_S12.sorted_stats |

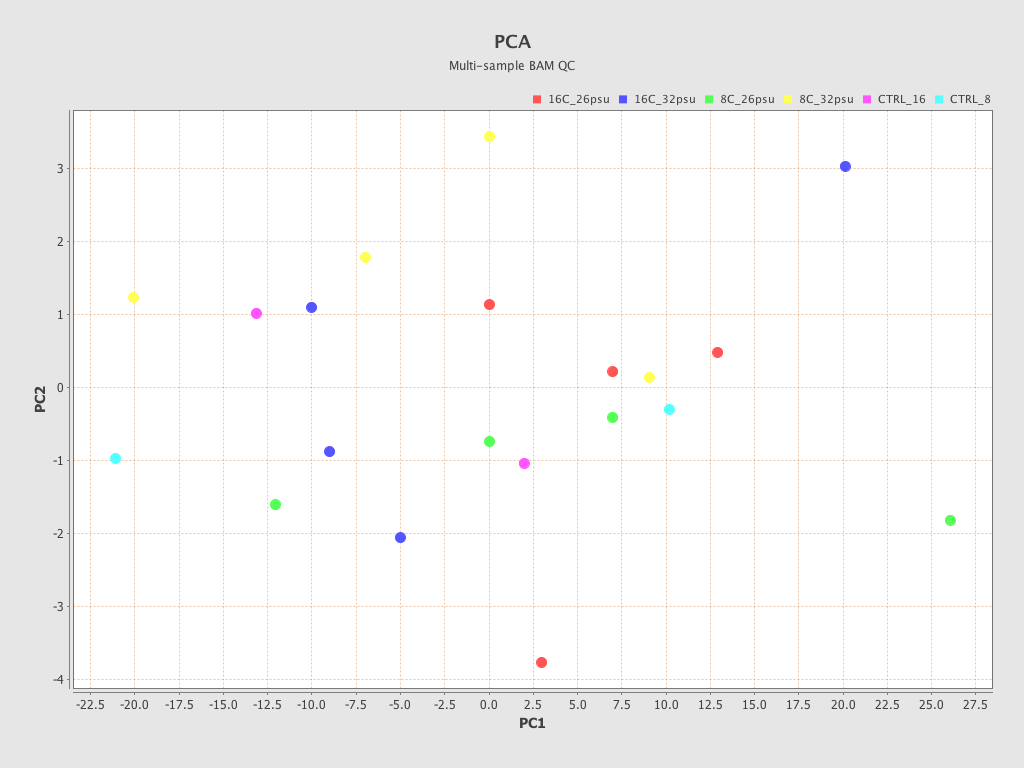
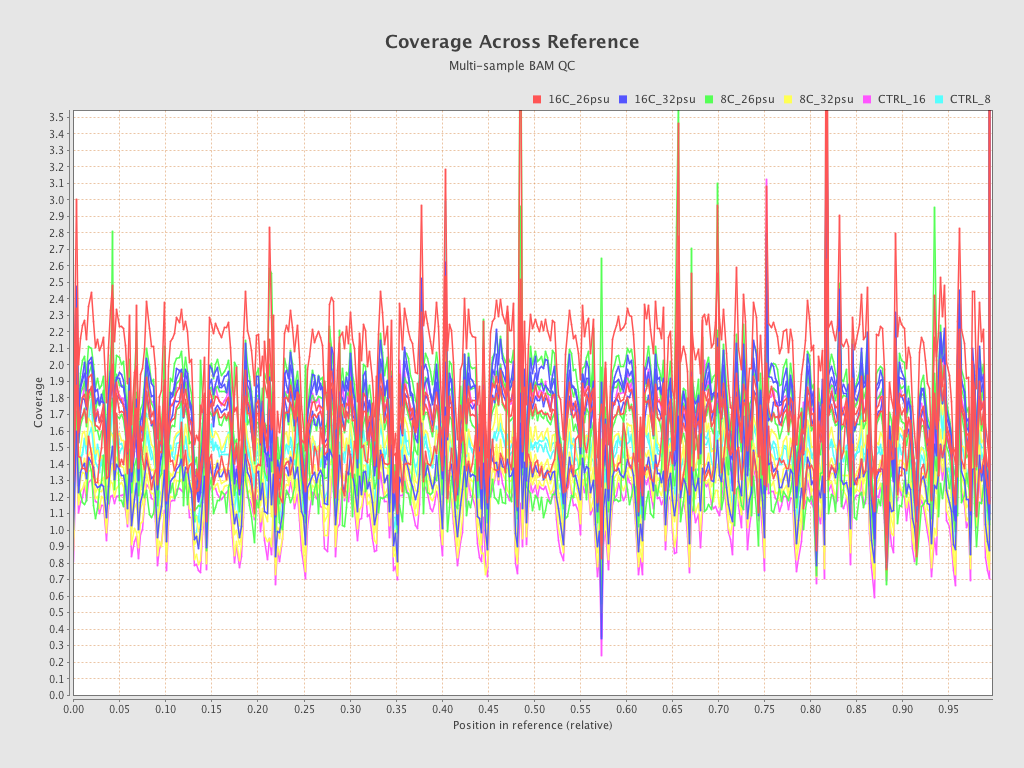
.png)