Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 28,751,448 |
| Mapped reads | 28,751,448 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 28,751,448 / 100% |
| Mapped reads, first in pair | 14,375,724 / 50% |
| Mapped reads, second in pair | 14,375,724 / 50% |
| Mapped reads, both in pair | 28,751,448 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 111.89 |
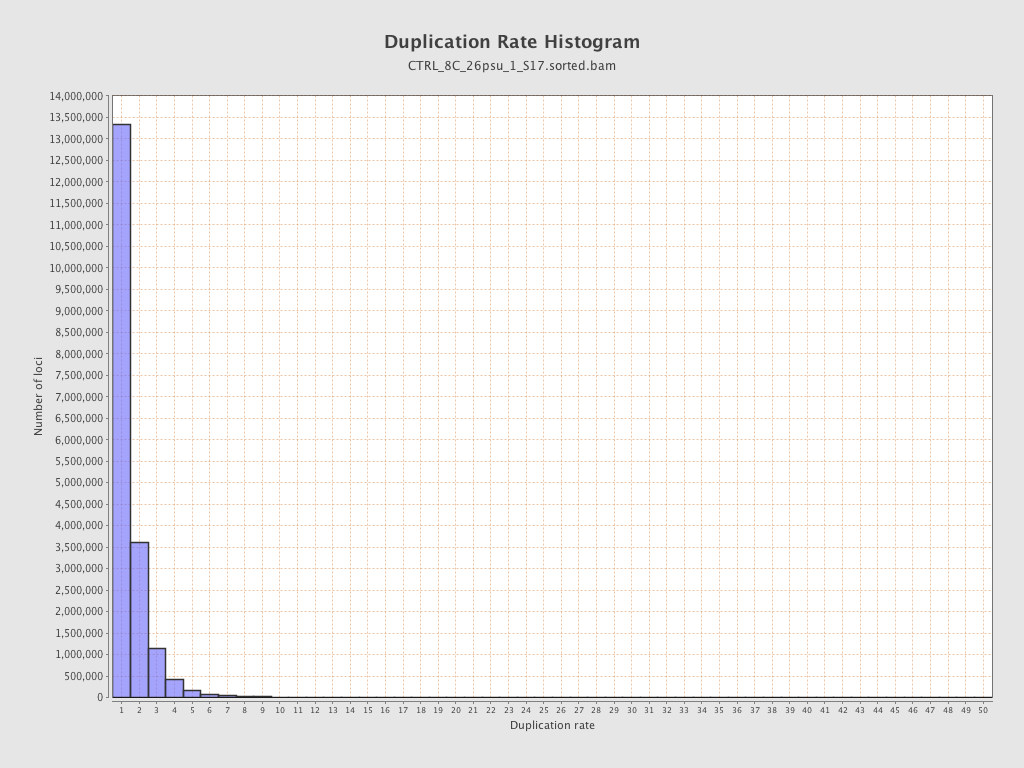
| Duplicated reads (estimated) | 9,871,737 / 34.33% |
| Duplication rate | 29.31% |
| Clipped reads | 0 / 0% |
ACGT Content
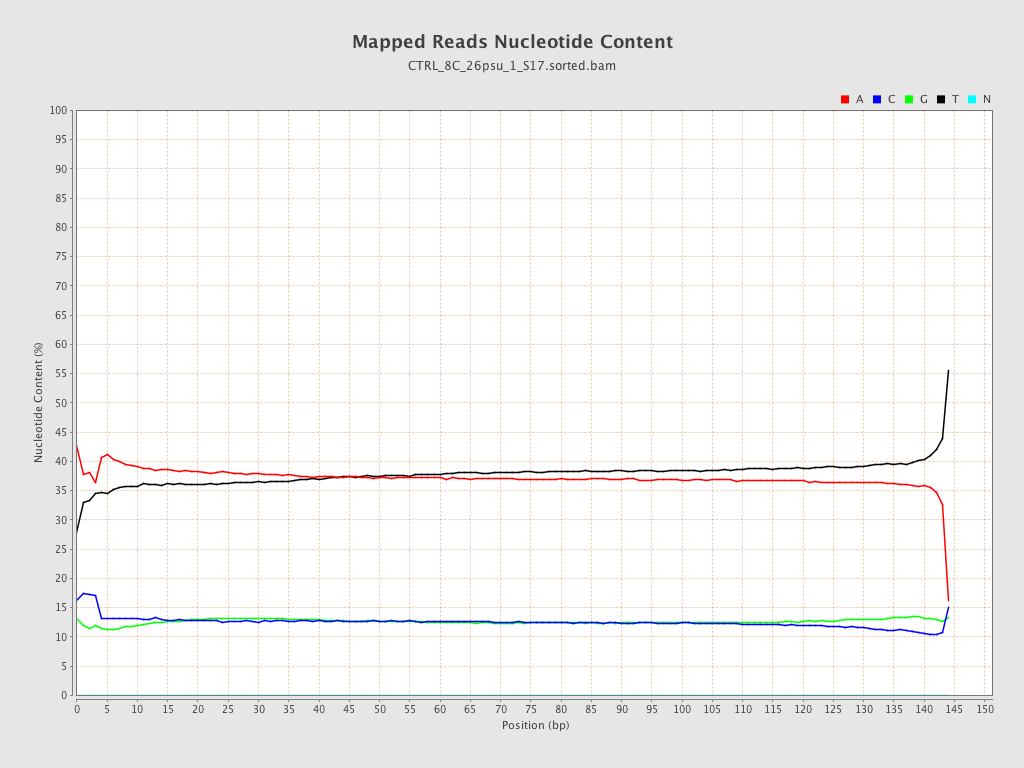
| Number/percentage of A's | 1,196,463,142 / 37.42% |
| Number/percentage of C's | 404,189,548 / 12.64% |
| Number/percentage of T's | 1,195,164,434 / 37.38% |
| Number/percentage of G's | 401,663,891 / 12.56% |
| Number/percentage of N's | 18,397 / 0% |
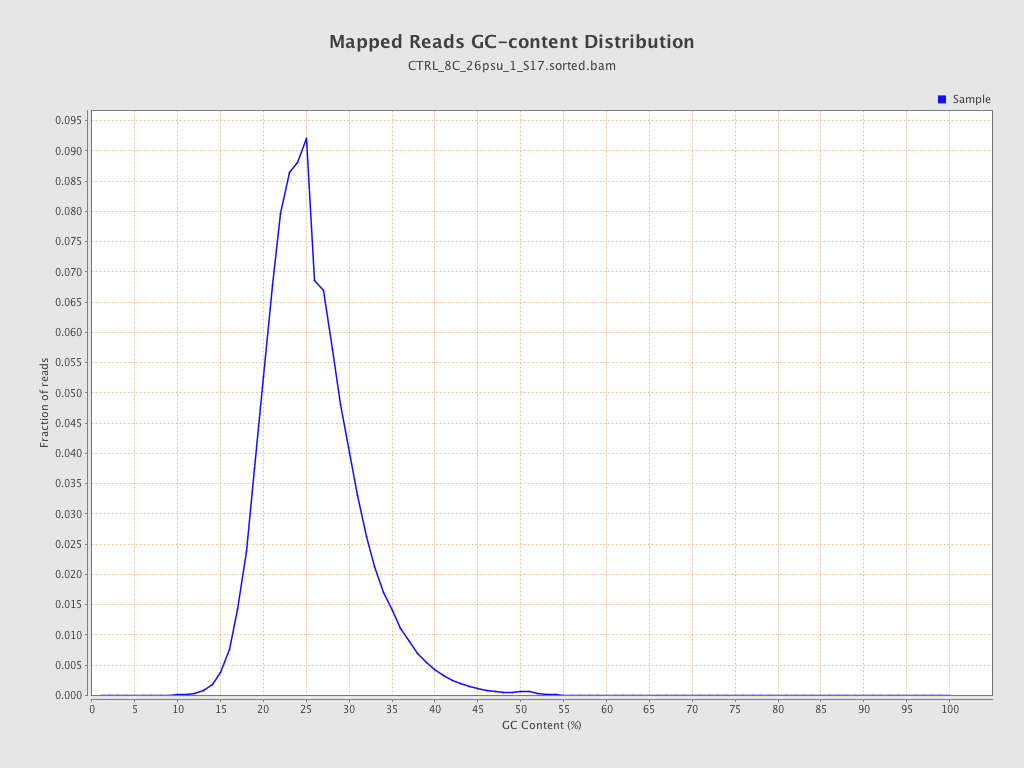
| GC Percentage | 25.2% |
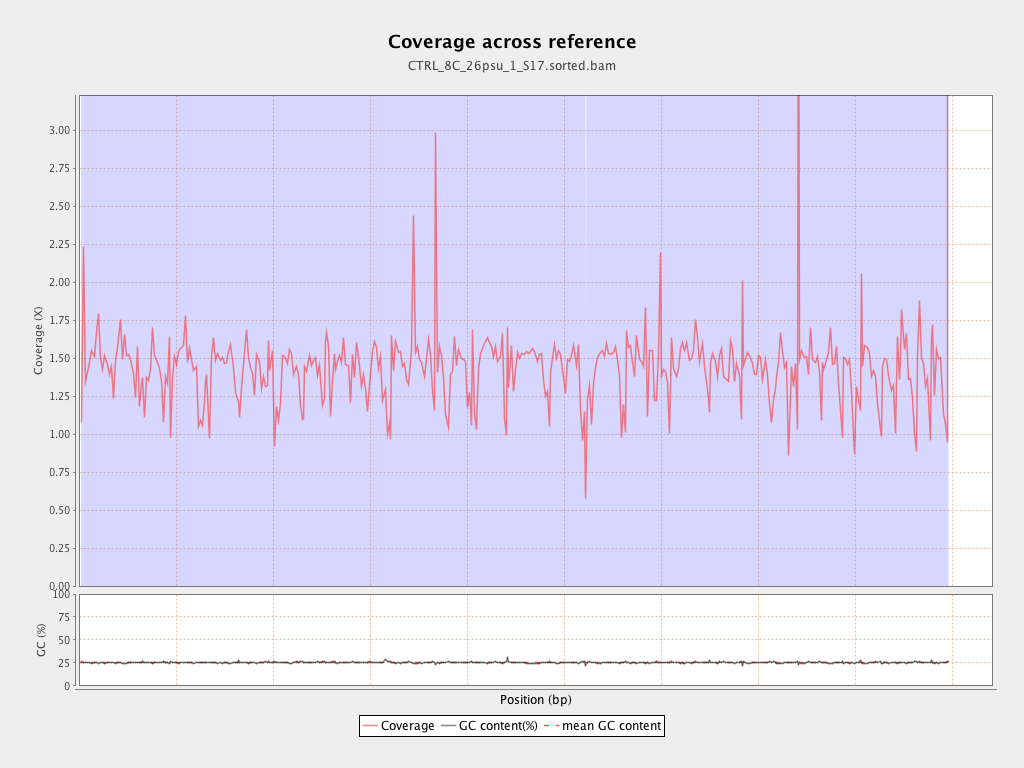
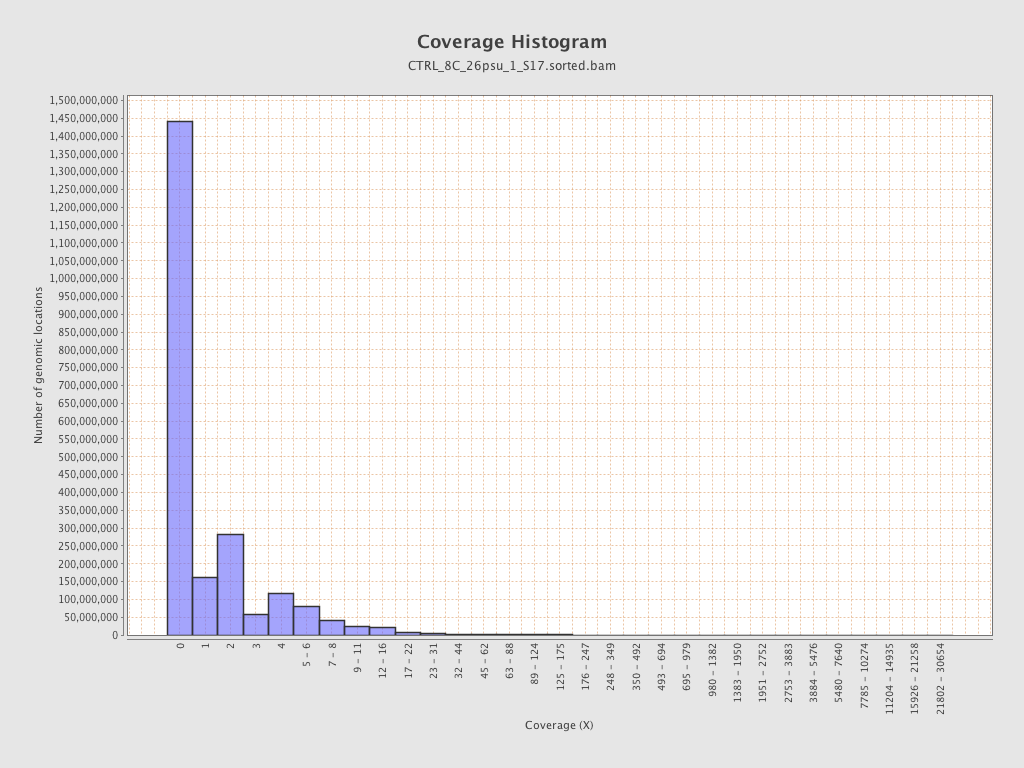
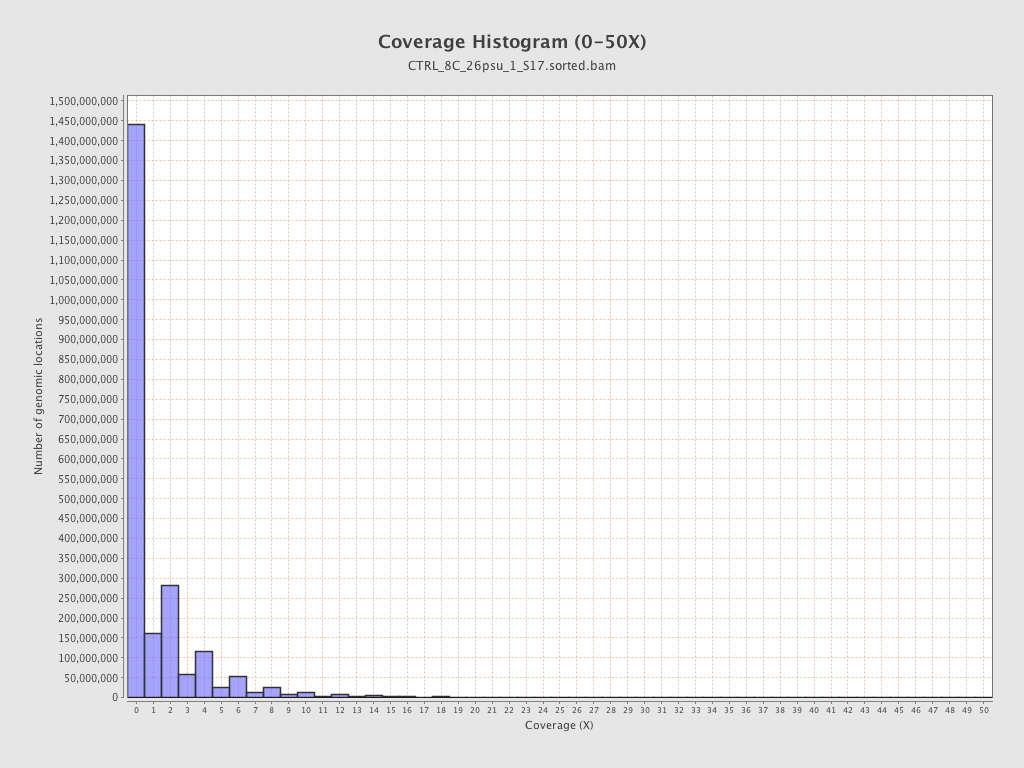
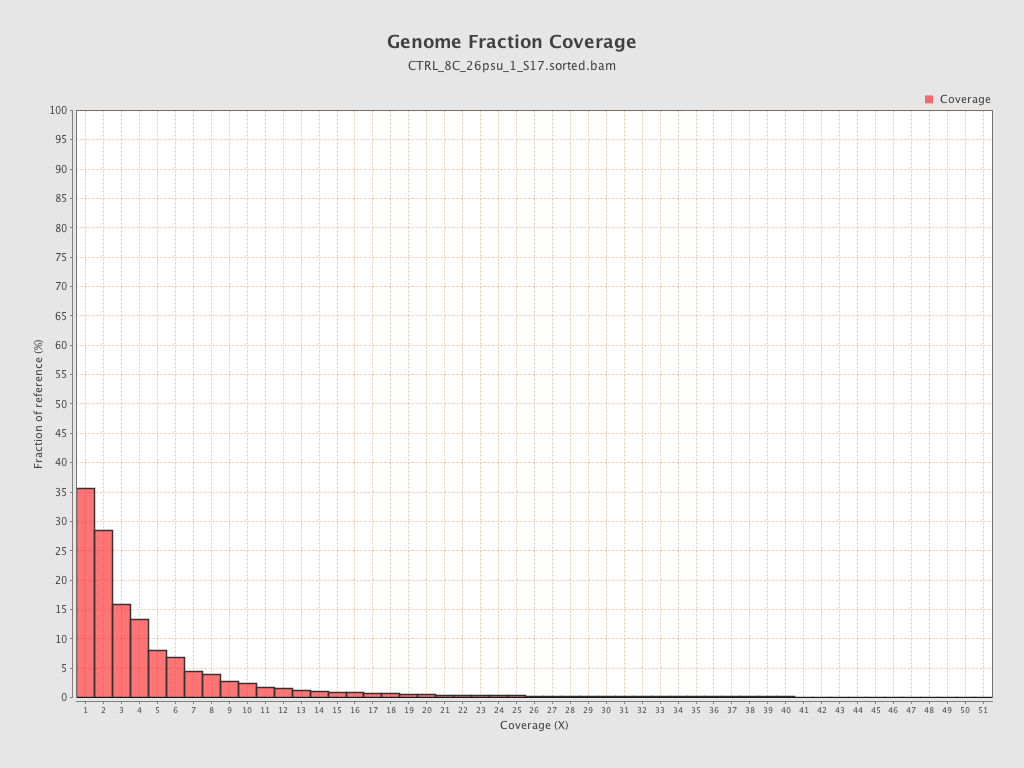
Coverage
| Mean | 1.4311 |
| Standard Deviation | 11.9724 |
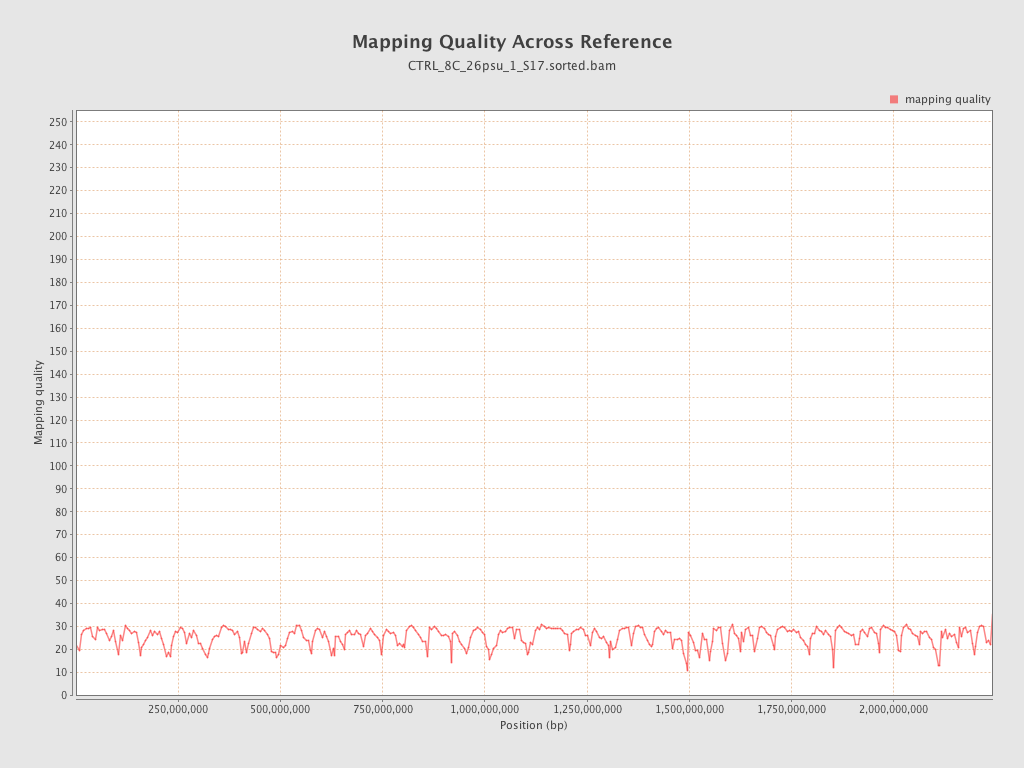
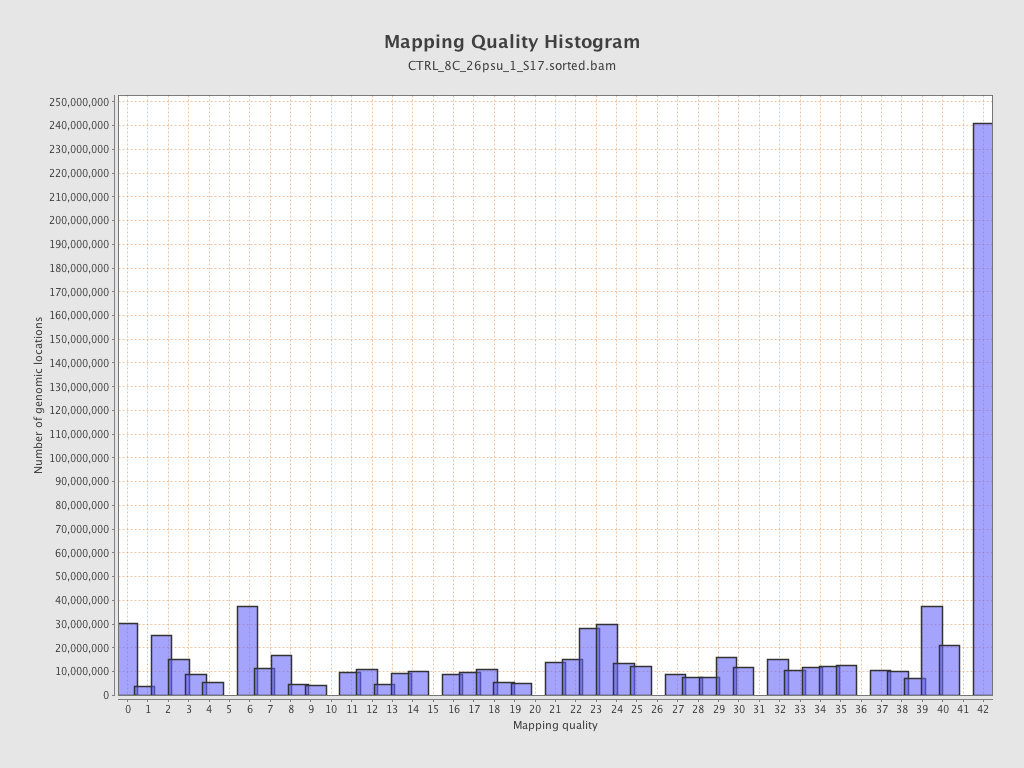
Mapping Quality
| Mean Mapping Quality | 25.31 |
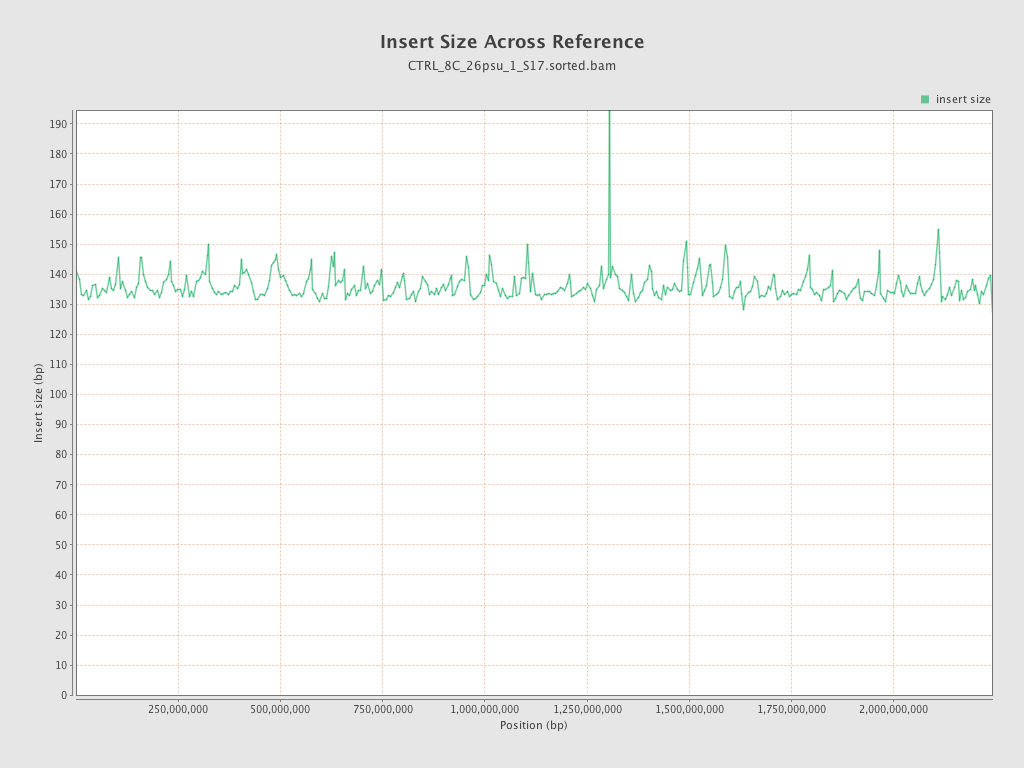
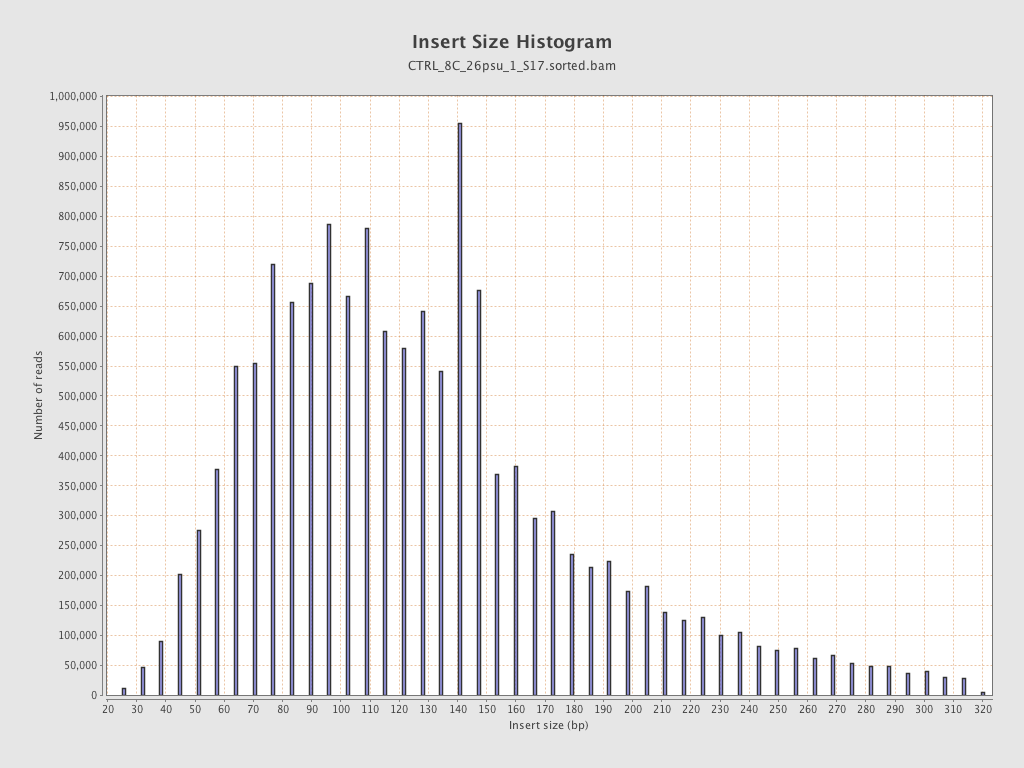
Insert size
| Mean | 136.04 |
| Standard Deviation | 66.66 |
| P25/Median/P75 | 90 / 123 / 160 |
Mismatches and indels
| General error rate | 20.2% |
| Mismatches | 627,433,268 |
| Insertions | 6,469,093 |
| Mapped reads with at least one insertion | 16.07% |
| Deletions | 5,172,131 |
| Mapped reads with at least one deletion | 15.45% |
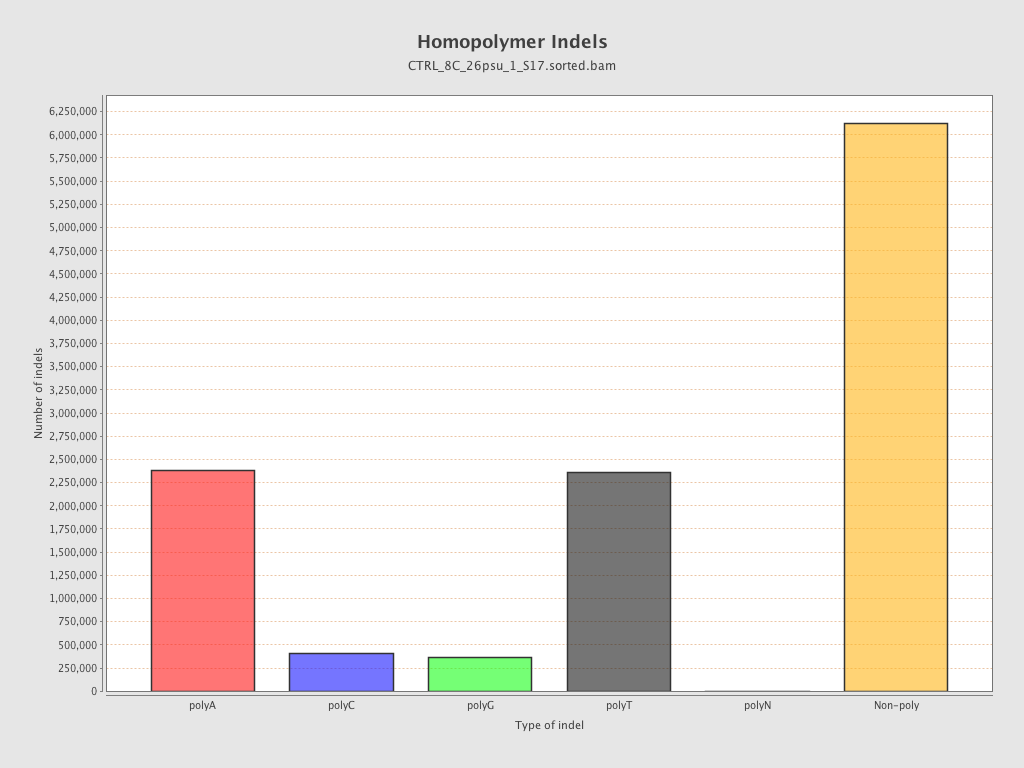
| Homopolymer indels | 47.43% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 237750512 | 1.4949 | 9.2787 |
| NC_027301.1 | 72943711 | 101120495 | 1.3863 | 7.9163 |
| NC_027302.1 | 92503428 | 129305799 | 1.3978 | 5.9124 |
| NC_027303.1 | 82398023 | 118746218 | 1.4411 | 5.7681 |
| NC_027304.1 | 80503876 | 111676864 | 1.3872 | 10.3524 |
| NC_027305.1 | 87043187 | 117275337 | 1.3473 | 8.3674 |
| NC_027306.1 | 58785265 | 82082718 | 1.3963 | 8.4952 |
| NC_027307.1 | 26434011 | 38148038 | 1.4431 | 8.886 |
| NC_027308.1 | 141712163 | 197995035 | 1.3972 | 6.2089 |
| NC_027309.1 | 116138521 | 176602101 | 1.5206 | 34.6464 |
| NC_027310.1 | 93888508 | 132009951 | 1.406 | 6.9138 |
| NC_027311.1 | 91880962 | 132405106 | 1.4411 | 7.1038 |
| NC_027312.1 | 107758822 | 157322974 | 1.46 | 8.1924 |
| NC_027313.1 | 93901823 | 133173694 | 1.4182 | 7.3984 |
| NC_027314.1 | 103963436 | 144952805 | 1.3943 | 4.6796 |
| NC_027315.1 | 87796322 | 130786916 | 1.4897 | 12.677 |
| NC_027316.1 | 57682537 | 81431033 | 1.4117 | 11.312 |
| NC_027317.1 | 70695409 | 106940548 | 1.5127 | 7.6202 |
| NC_027318.1 | 82978132 | 118946455 | 1.4335 | 12.8525 |
| NC_027319.1 | 86795997 | 121432659 | 1.3991 | 5.6394 |
| NC_027320.1 | 58021487 | 79862922 | 1.3764 | 5.6663 |
| NC_027321.1 | 63420196 | 96232151 | 1.5174 | 14.3766 |
| NC_027322.1 | 49854004 | 72094425 | 1.4461 | 8.0125 |
| NC_027323.1 | 48650976 | 63909730 | 1.3136 | 5.0834 |
| NC_027324.1 | 51481326 | 72824735 | 1.4146 | 5.9001 |
| NC_027325.1 | 47900953 | 66144280 | 1.3809 | 7.93 |
| NC_027326.1 | 43943985 | 62016205 | 1.4113 | 9.0726 |
| NC_027327.1 | 39600944 | 56896326 | 1.4367 | 10.3346 |
| NC_027328.1 | 42488238 | 55828070 | 1.314 | 12.4619 |
| NC_001960.1 | 16665 | 10147196 | 608.8926 | 1,103.789 |