Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 25,716,104 |
| Mapped reads | 25,716,104 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 25,716,104 / 100% |
| Mapped reads, first in pair | 12,858,052 / 50% |
| Mapped reads, second in pair | 12,858,052 / 50% |
| Mapped reads, both in pair | 25,716,104 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 97.77 |
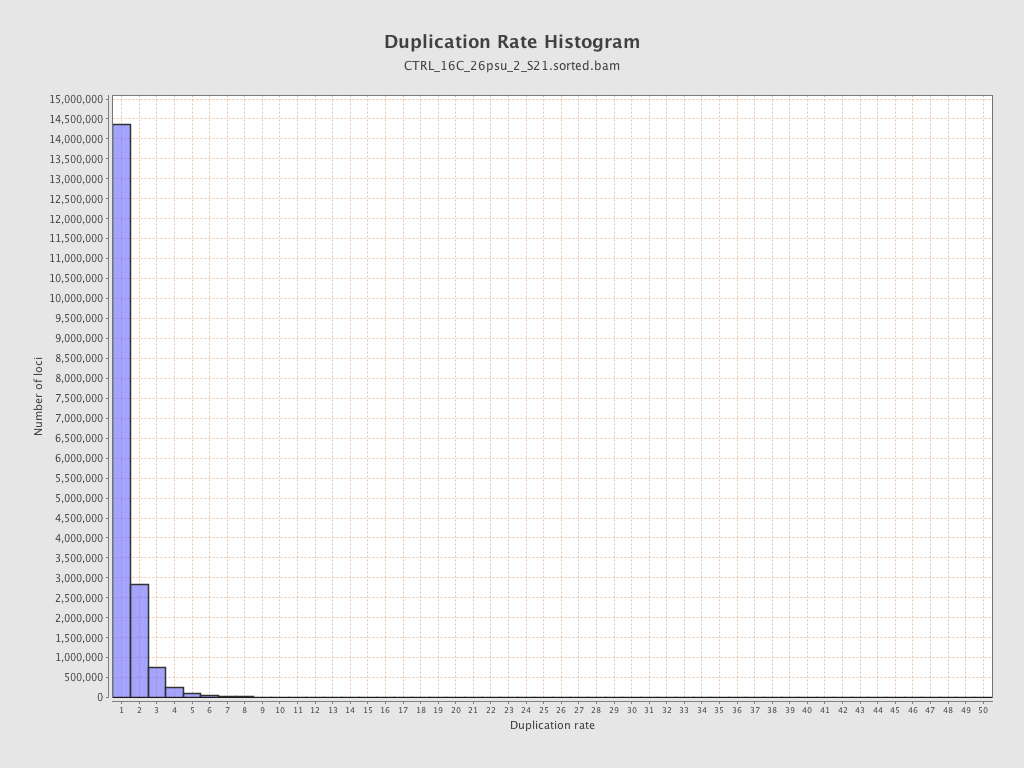
| Duplicated reads (estimated) | 7,218,919 / 28.07% |
| Duplication rate | 22.29% |
| Clipped reads | 0 / 0% |
ACGT Content
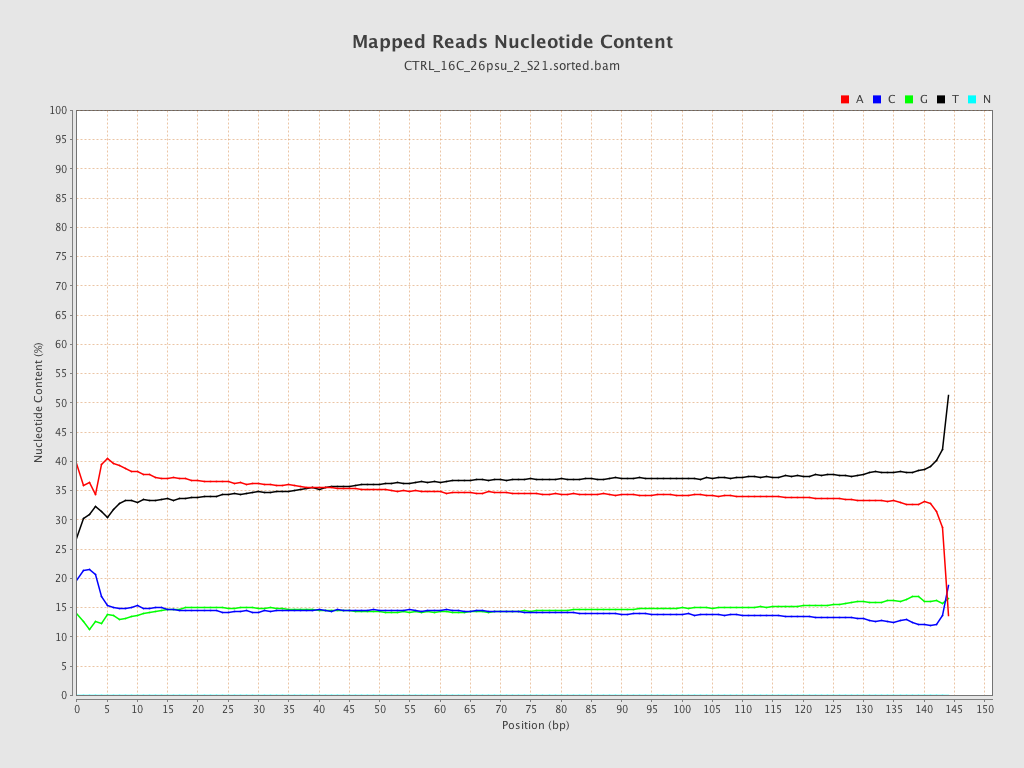
| Number/percentage of A's | 885,166,319 / 35.52% |
| Number/percentage of C's | 363,228,368 / 14.58% |
| Number/percentage of T's | 883,372,273 / 35.45% |
| Number/percentage of G's | 360,291,314 / 14.46% |
| Number/percentage of N's | 14,647 / 0% |
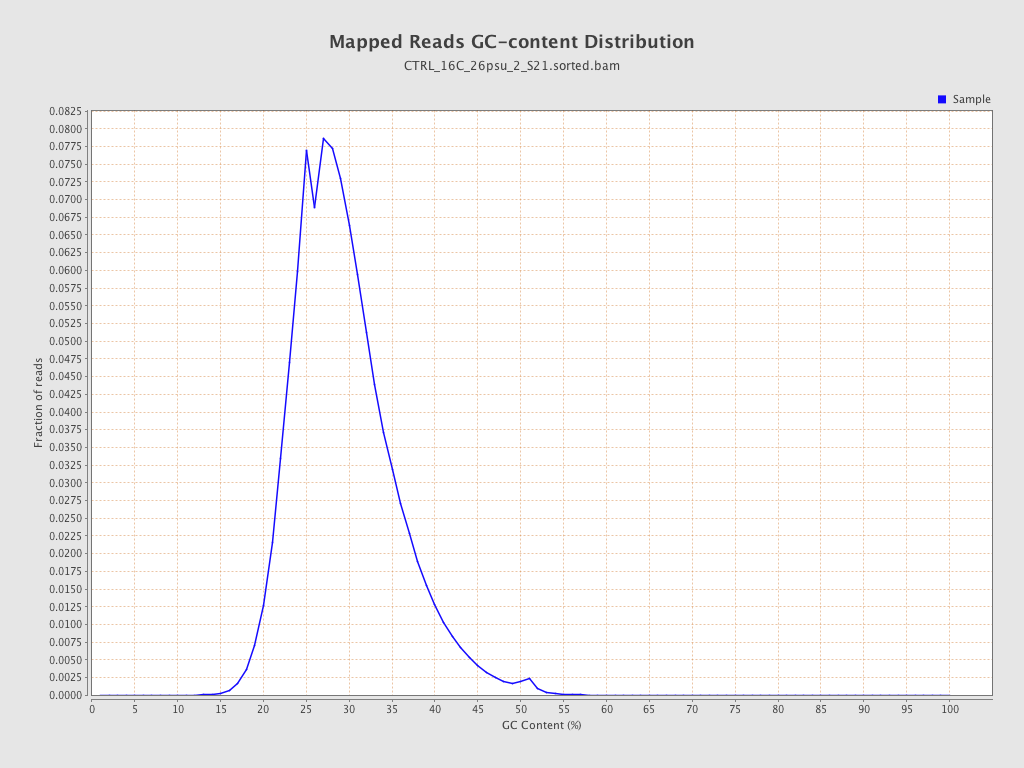
| GC Percentage | 29.03% |
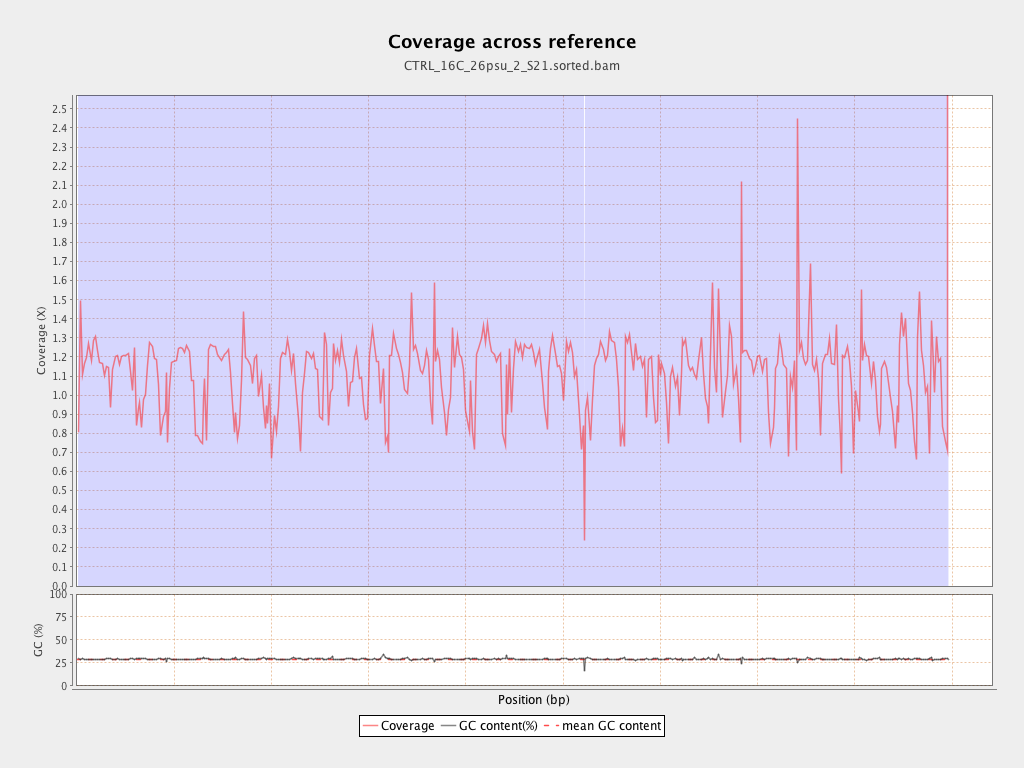
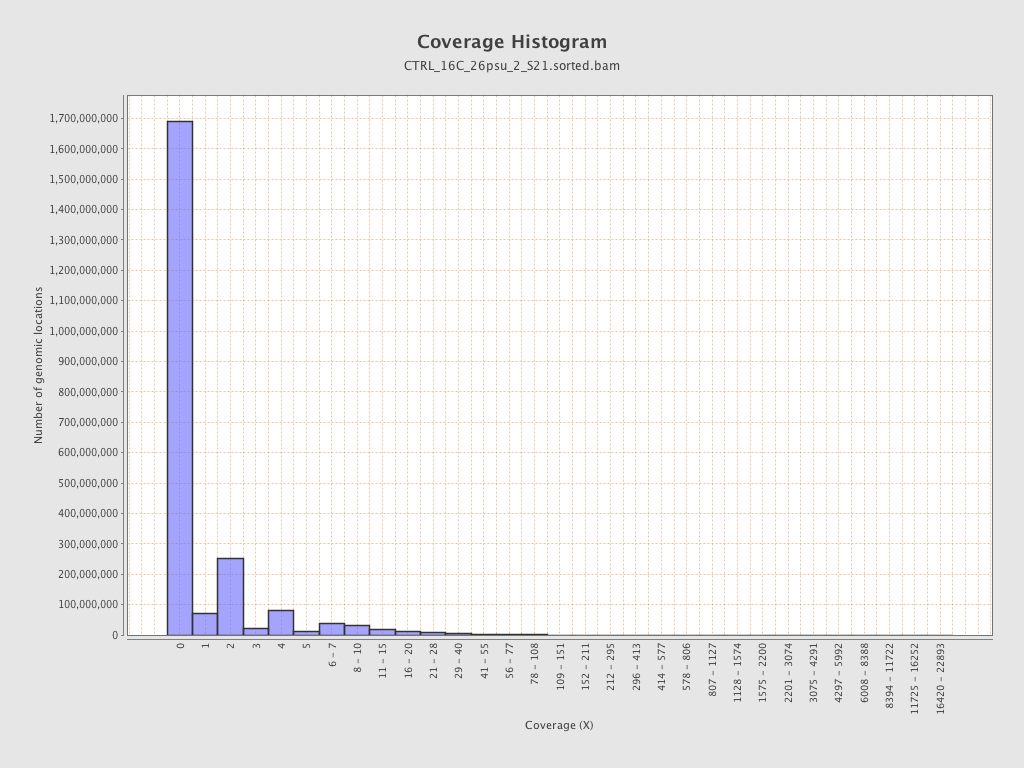
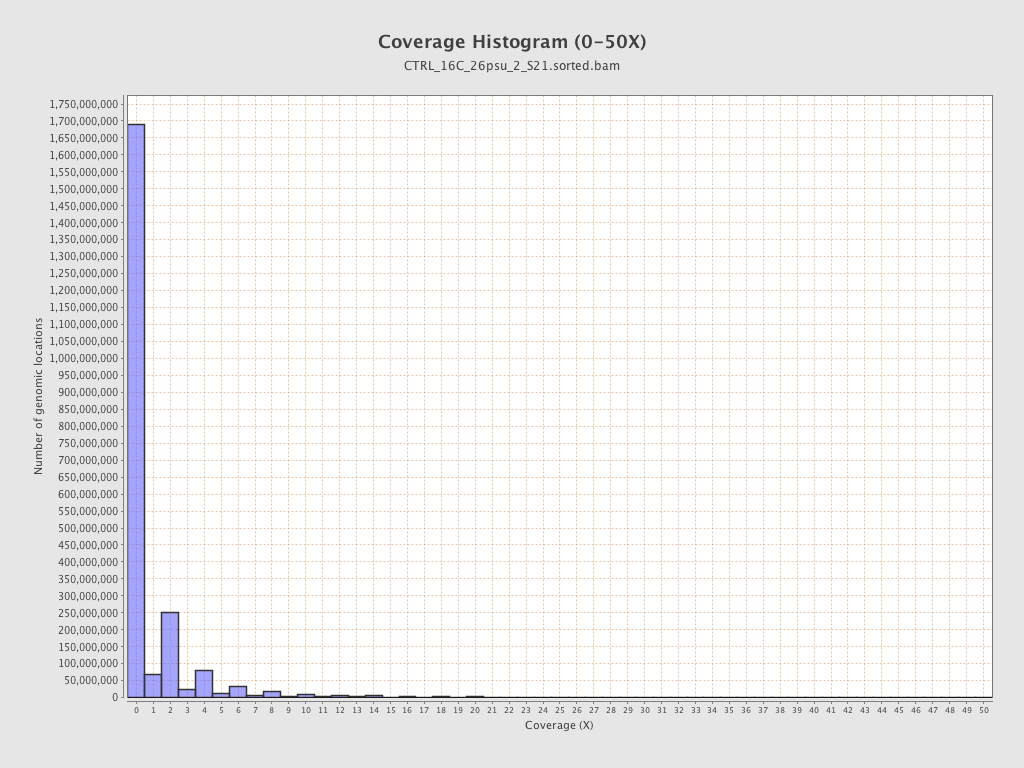
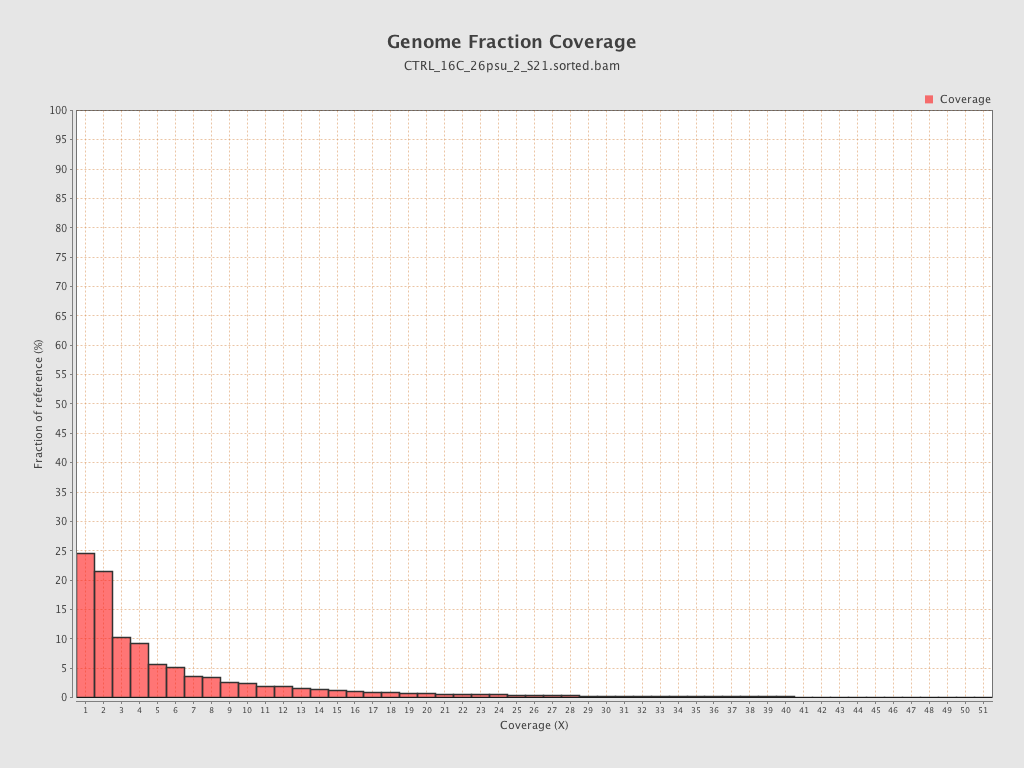
Coverage
| Mean | 1.1158 |
| Standard Deviation | 10.7513 |
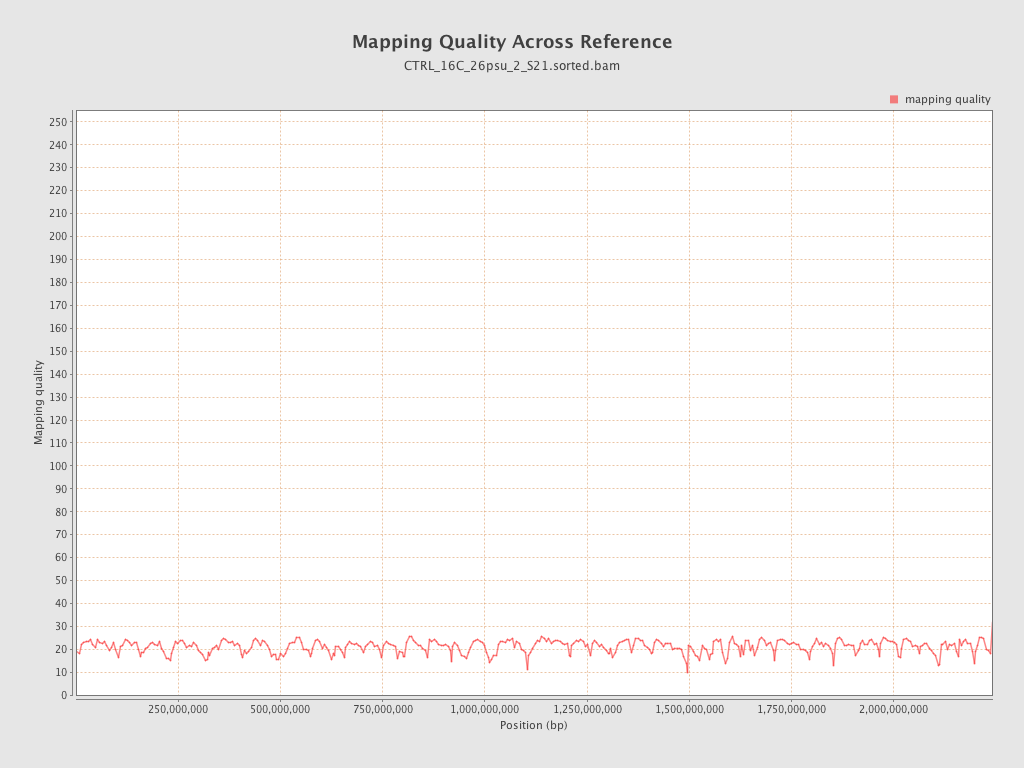
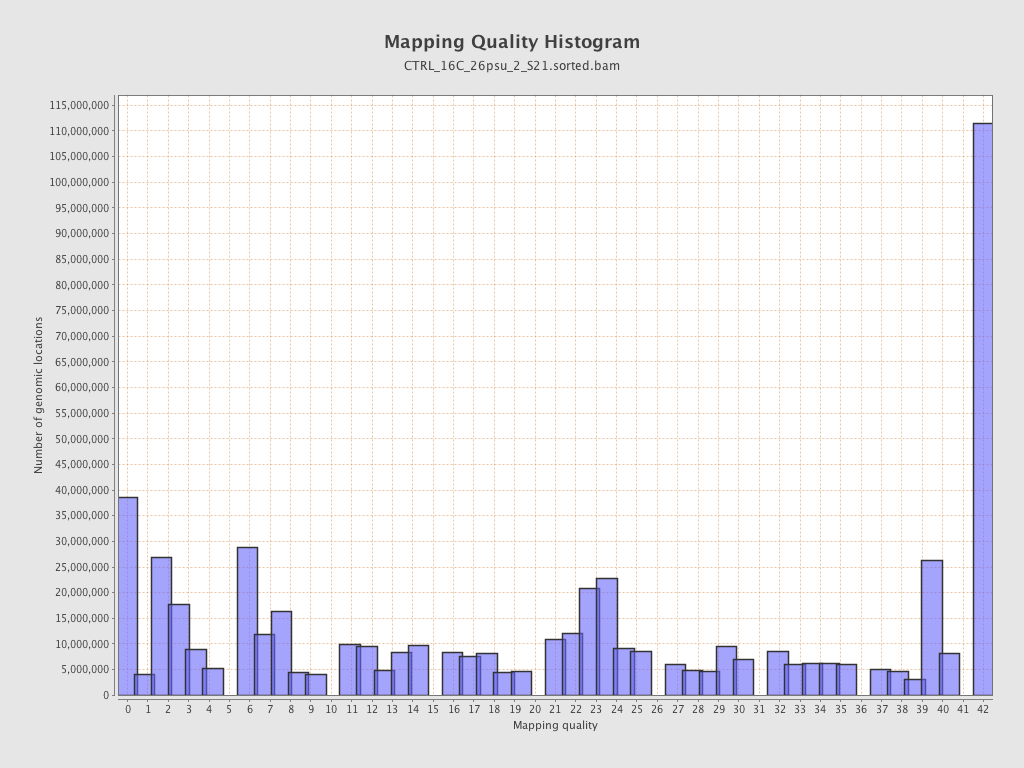
Mapping Quality
| Mean Mapping Quality | 21.08 |
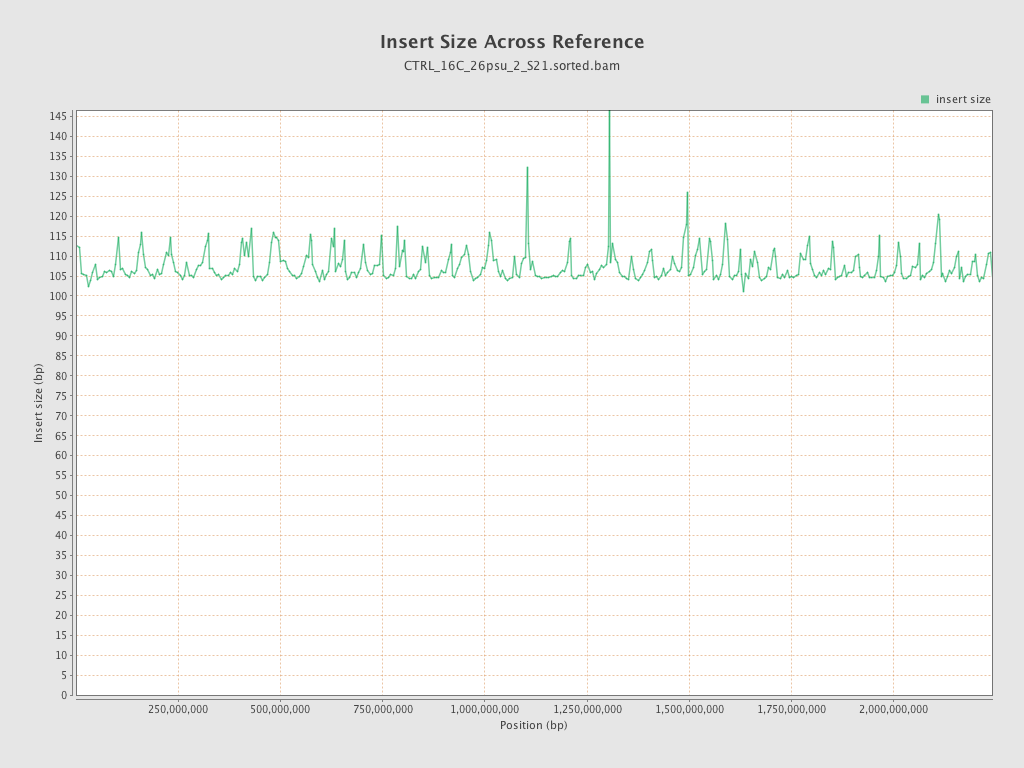
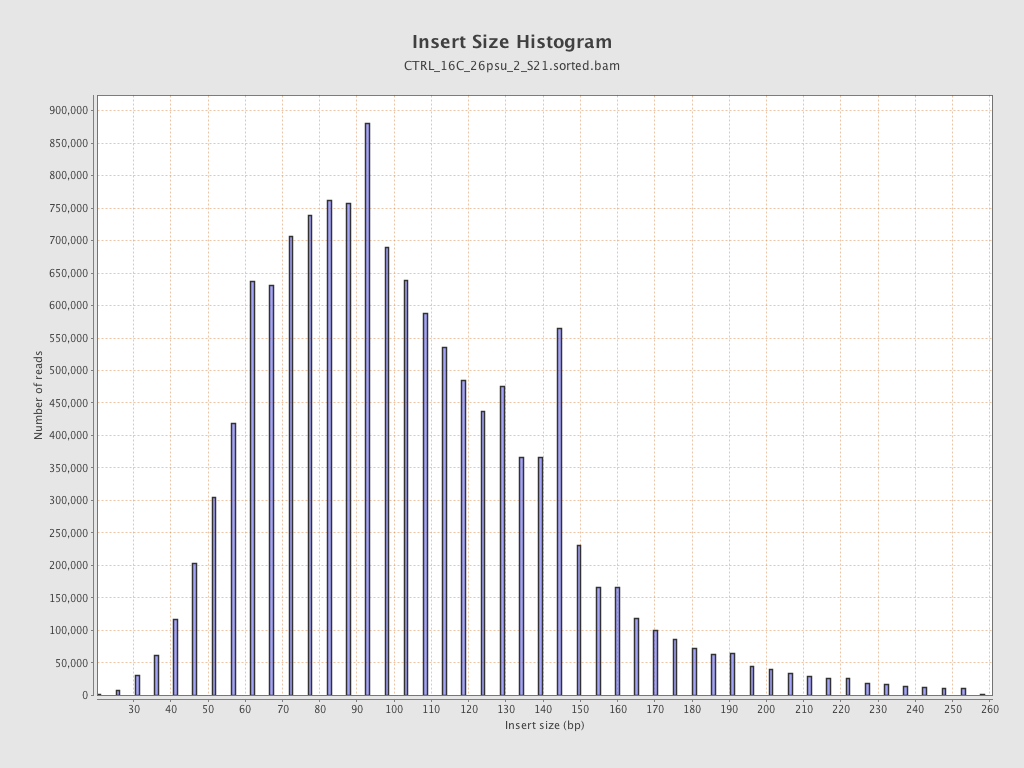
Insert size
| Mean | 107.18 |
| Standard Deviation | 43.02 |
| P25/Median/P75 | 78 / 100 / 129 |
Mismatches and indels
| General error rate | 21.66% |
| Mismatches | 518,329,887 |
| Insertions | 7,546,269 |
| Mapped reads with at least one insertion | 21.56% |
| Deletions | 4,686,411 |
| Mapped reads with at least one deletion | 15.69% |
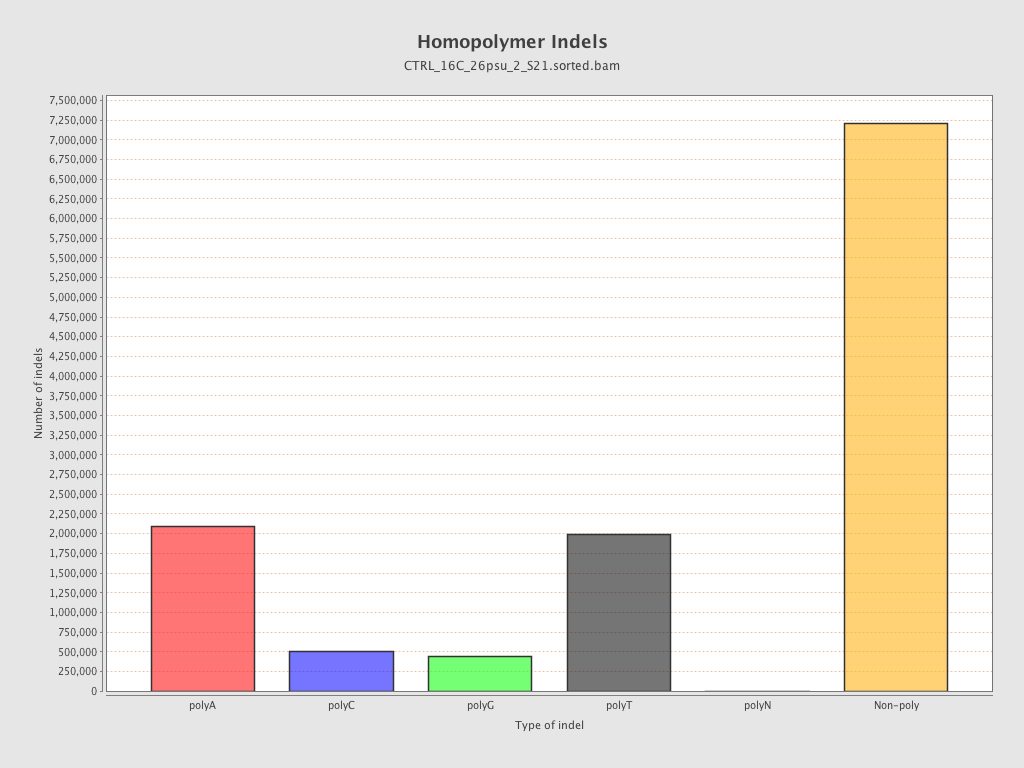
| Homopolymer indels | 41.11% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 184107560 | 1.1576 | 5.8099 |
| NC_027301.1 | 72943711 | 75988043 | 1.0417 | 4.5314 |
| NC_027302.1 | 92503428 | 98811669 | 1.0682 | 4.0757 |
| NC_027303.1 | 82398023 | 94013586 | 1.141 | 9.0555 |
| NC_027304.1 | 80503876 | 85847933 | 1.0664 | 6.5113 |
| NC_027305.1 | 87043187 | 90096321 | 1.0351 | 7.4431 |
| NC_027306.1 | 58785265 | 63837548 | 1.0859 | 5.8281 |
| NC_027307.1 | 26434011 | 28988500 | 1.0966 | 9.6977 |
| NC_027308.1 | 141712163 | 155336762 | 1.0961 | 5.63 |
| NC_027309.1 | 116138521 | 136149232 | 1.1723 | 13.58 |
| NC_027310.1 | 93888508 | 101365686 | 1.0796 | 5.3247 |
| NC_027311.1 | 91880962 | 104142033 | 1.1334 | 8.9942 |
| NC_027312.1 | 107758822 | 123676598 | 1.1477 | 5.8249 |
| NC_027313.1 | 93901823 | 104895661 | 1.1171 | 8.0551 |
| NC_027314.1 | 103963436 | 113122739 | 1.0881 | 4.5163 |
| NC_027315.1 | 87796322 | 99646566 | 1.135 | 6.0173 |
| NC_027316.1 | 57682537 | 59918835 | 1.0388 | 5.1128 |
| NC_027317.1 | 70695409 | 80854496 | 1.1437 | 8.1968 |
| NC_027318.1 | 82978132 | 95964223 | 1.1565 | 29.9279 |
| NC_027319.1 | 86795997 | 96466545 | 1.1114 | 5.5928 |
| NC_027320.1 | 58021487 | 62702866 | 1.0807 | 4.0107 |
| NC_027321.1 | 63420196 | 76597573 | 1.2078 | 19.897 |
| NC_027322.1 | 49854004 | 57121376 | 1.1458 | 5.7108 |
| NC_027323.1 | 48650976 | 50921091 | 1.0467 | 4.3205 |
| NC_027324.1 | 51481326 | 57192841 | 1.1109 | 5.5842 |
| NC_027325.1 | 47900953 | 47959267 | 1.0012 | 5.3068 |
| NC_027326.1 | 43943985 | 49023628 | 1.1156 | 6.1874 |
| NC_027327.1 | 39600944 | 45449119 | 1.1477 | 7.9729 |
| NC_027328.1 | 42488238 | 43364173 | 1.0206 | 6.6231 |
| NC_001960.1 | 16665 | 16023712 | 961.5189 | 1,622.0133 |