Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 34,900,622 |
| Mapped reads | 34,900,622 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 34,900,622 / 100% |
| Mapped reads, first in pair | 17,450,311 / 50% |
| Mapped reads, second in pair | 17,450,311 / 50% |
| Mapped reads, both in pair | 34,900,622 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 108.1 |
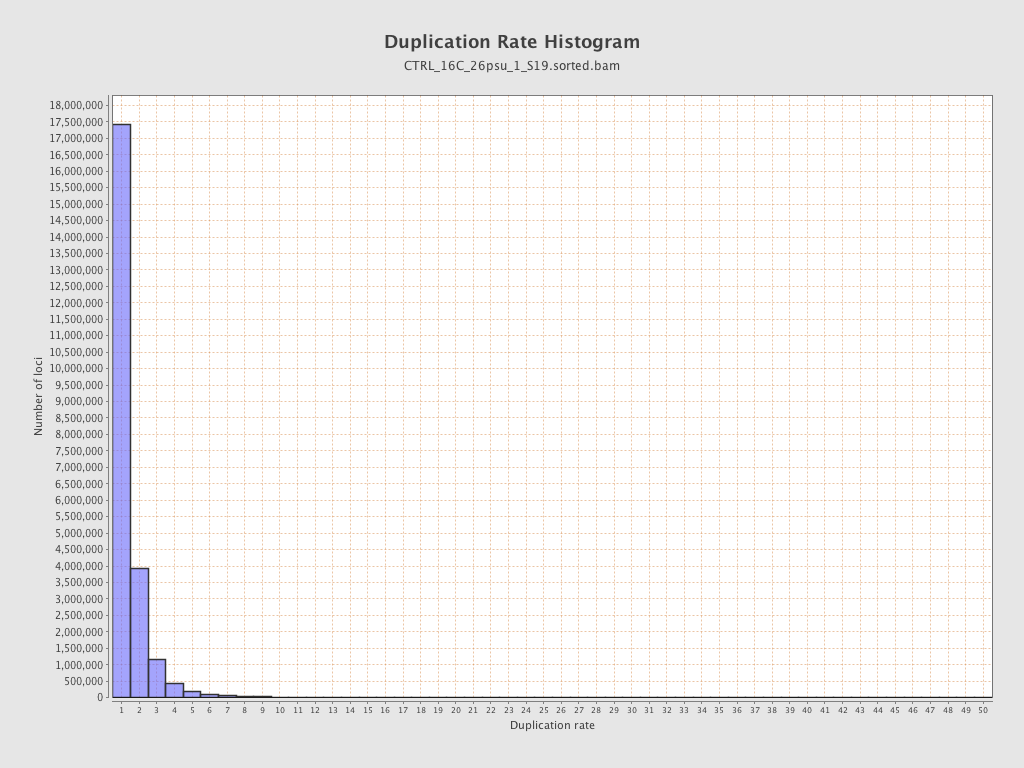
| Duplicated reads (estimated) | 11,424,033 / 32.73% |
| Duplication rate | 25.74% |
| Clipped reads | 0 / 0% |
ACGT Content
| Number/percentage of A's | 1,360,794,773 / 36.35% |
| Number/percentage of C's | 511,864,593 / 13.67% |
| Number/percentage of T's | 1,360,169,007 / 36.34% |
| Number/percentage of G's | 510,533,696 / 13.64% |
| Number/percentage of N's | 23,982 / 0% |
| GC Percentage | 27.31% |
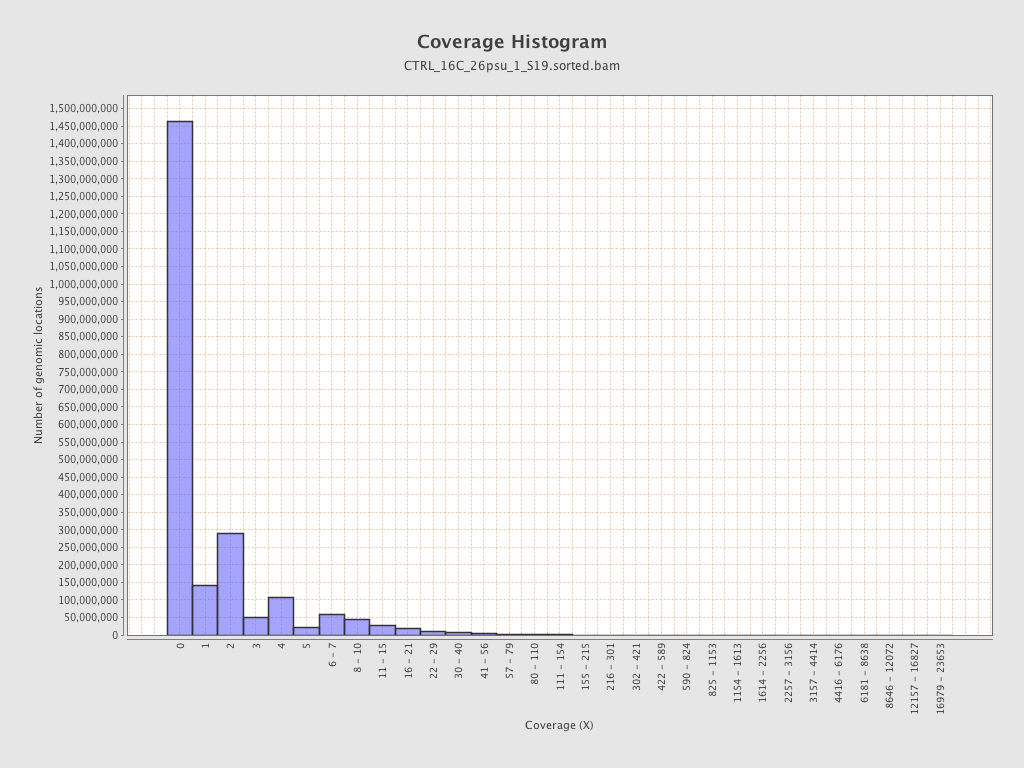
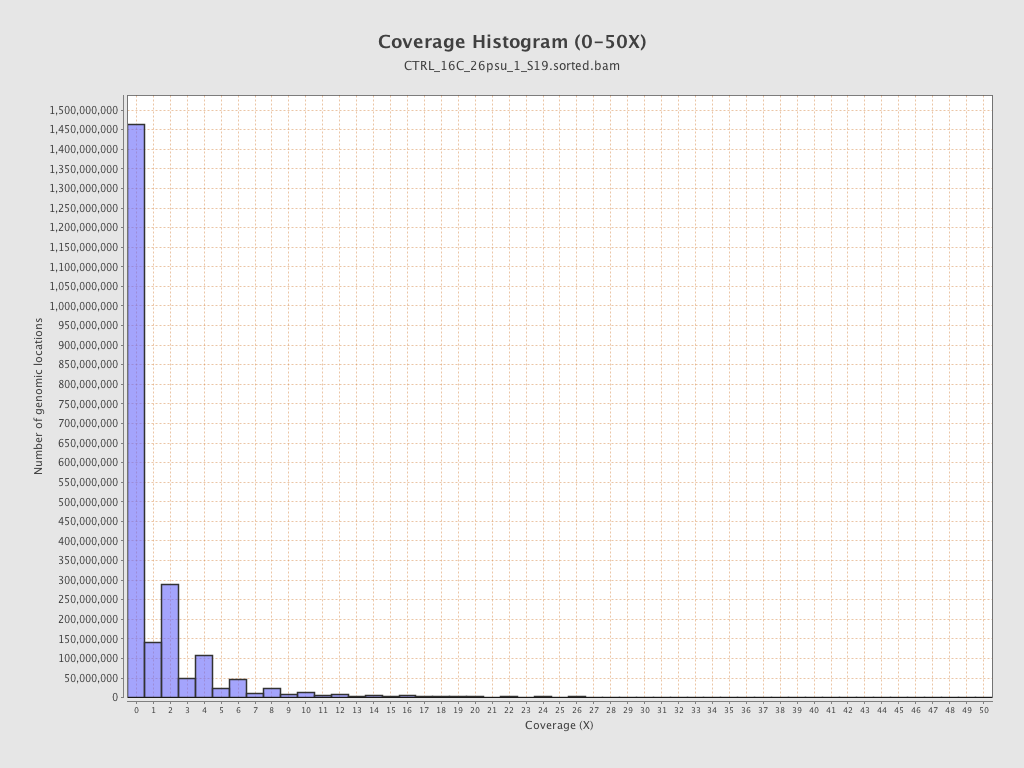
Coverage
| Mean | 1.6756 |
| Standard Deviation | 12.7065 |
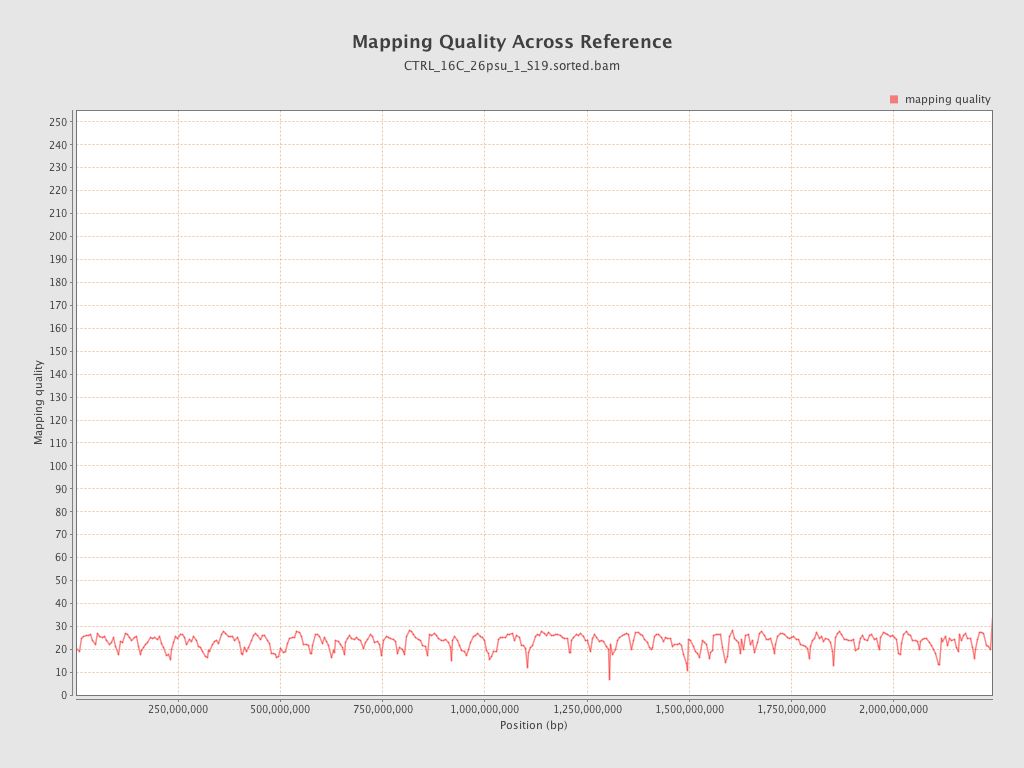
Mapping Quality
| Mean Mapping Quality | 23.11 |
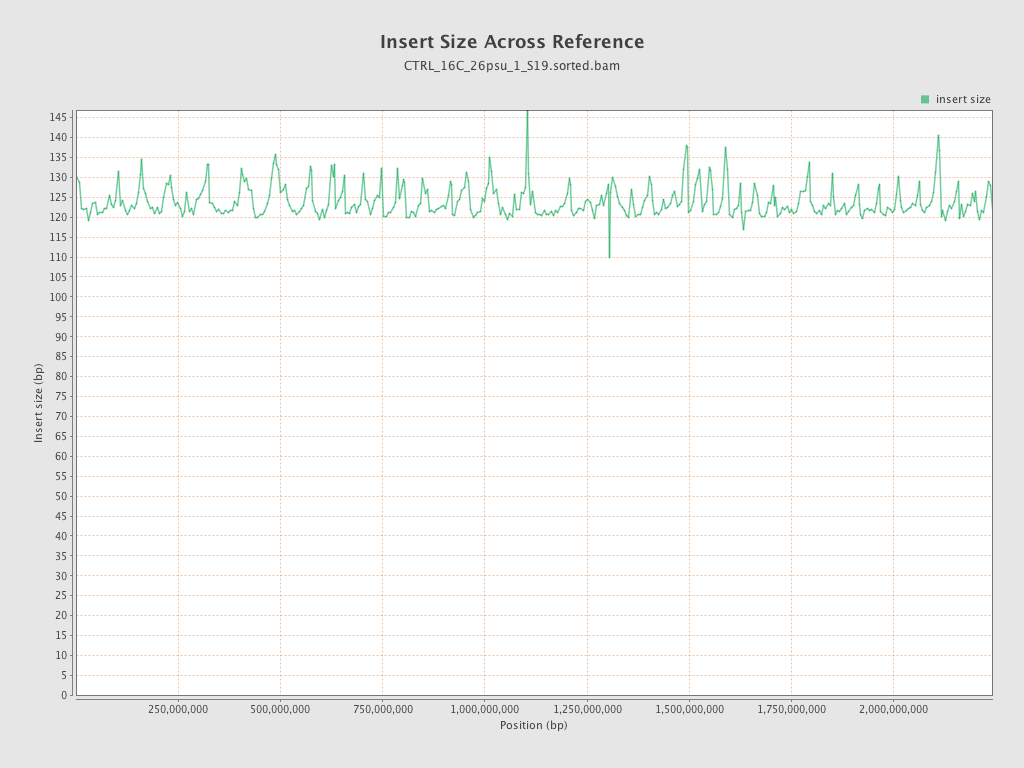
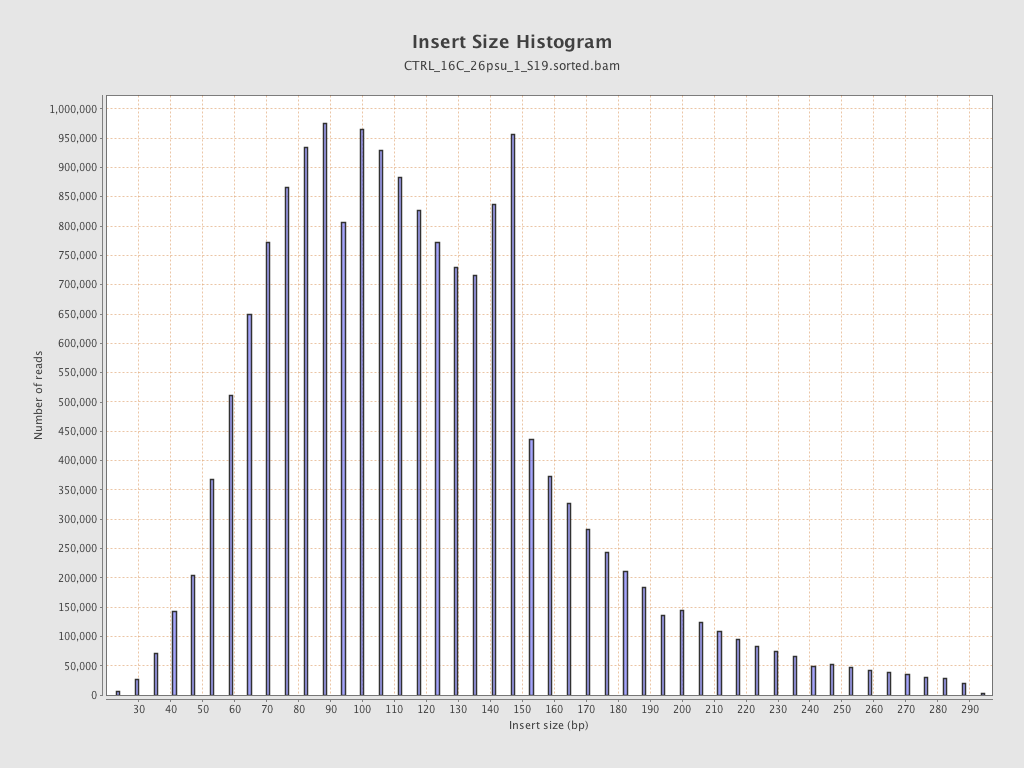
Insert size
| Mean | 124 |
| Standard Deviation | 54.57 |
| P25/Median/P75 | 87 / 115 / 147 |
Mismatches and indels
| General error rate | 21.26% |
| Mismatches | 767,196,564 |
| Insertions | 9,631,319 |
| Mapped reads with at least one insertion | 19.26% |
| Deletions | 6,391,426 |
| Mapped reads with at least one deletion | 15.6% |
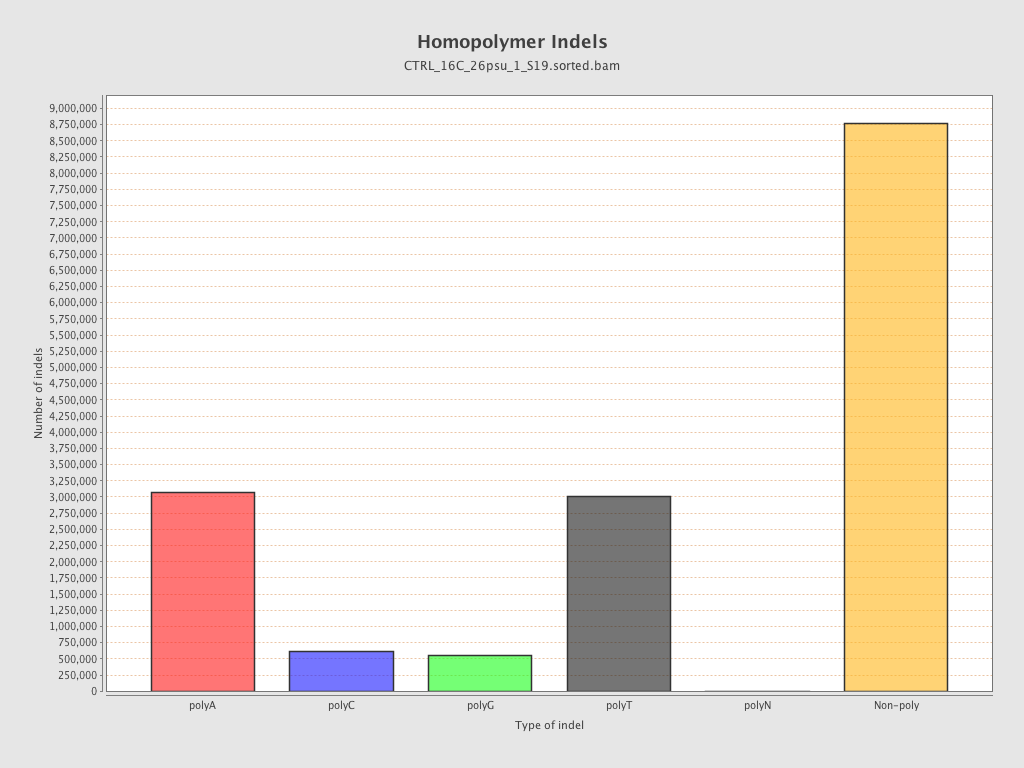
| Homopolymer indels | 45.31% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 277840406 | 1.747 | 9.1193 |
| NC_027301.1 | 72943711 | 116134203 | 1.5921 | 7.0855 |
| NC_027302.1 | 92503428 | 150488758 | 1.6268 | 5.8966 |
| NC_027303.1 | 82398023 | 140161012 | 1.701 | 9.5124 |
| NC_027304.1 | 80503876 | 129420116 | 1.6076 | 9.1768 |
| NC_027305.1 | 87043187 | 137887832 | 1.5841 | 14.3663 |
| NC_027306.1 | 58785265 | 96417077 | 1.6402 | 8.5985 |
| NC_027307.1 | 26434011 | 43908678 | 1.6611 | 13.4077 |
| NC_027308.1 | 141712163 | 234208645 | 1.6527 | 7.6349 |
| NC_027309.1 | 116138521 | 206221960 | 1.7757 | 26.551 |
| NC_027310.1 | 93888508 | 153726935 | 1.6373 | 7.1245 |
| NC_027311.1 | 91880962 | 155487080 | 1.6923 | 10.0155 |
| NC_027312.1 | 107758822 | 185139462 | 1.7181 | 7.951 |
| NC_027313.1 | 93901823 | 157557090 | 1.6779 | 7.9544 |
| NC_027314.1 | 103963436 | 170586098 | 1.6408 | 6.1965 |
| NC_027315.1 | 87796322 | 151919000 | 1.7304 | 10.1261 |
| NC_027316.1 | 57682537 | 92661461 | 1.6064 | 9.1762 |
| NC_027317.1 | 70695409 | 122240624 | 1.7291 | 10.6009 |
| NC_027318.1 | 82978132 | 142278714 | 1.7147 | 28.1739 |
| NC_027319.1 | 86795997 | 145698791 | 1.6786 | 7.7046 |
| NC_027320.1 | 58021487 | 94290514 | 1.6251 | 5.9335 |
| NC_027321.1 | 63420196 | 114456303 | 1.8047 | 25.3159 |
| NC_027322.1 | 49854004 | 85734400 | 1.7197 | 9.6121 |
| NC_027323.1 | 48650976 | 75939707 | 1.5609 | 6.0602 |
| NC_027324.1 | 51481326 | 85332153 | 1.6575 | 7.6134 |
| NC_027325.1 | 47900953 | 74163454 | 1.5483 | 7.5325 |
| NC_027326.1 | 43943985 | 73230042 | 1.6664 | 8.614 |
| NC_027327.1 | 39600944 | 66581611 | 1.6813 | 11.2374 |
| NC_027328.1 | 42488238 | 65606186 | 1.5441 | 12.7333 |
| NC_001960.1 | 16665 | 8496429 | 509.8367 | 862.0903 |