Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 28,602,080 |
| Mapped reads | 28,602,080 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 28,602,080 / 100% |
| Mapped reads, first in pair | 14,301,040 / 50% |
| Mapped reads, second in pair | 14,301,040 / 50% |
| Mapped reads, both in pair | 28,602,080 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 112.71 |
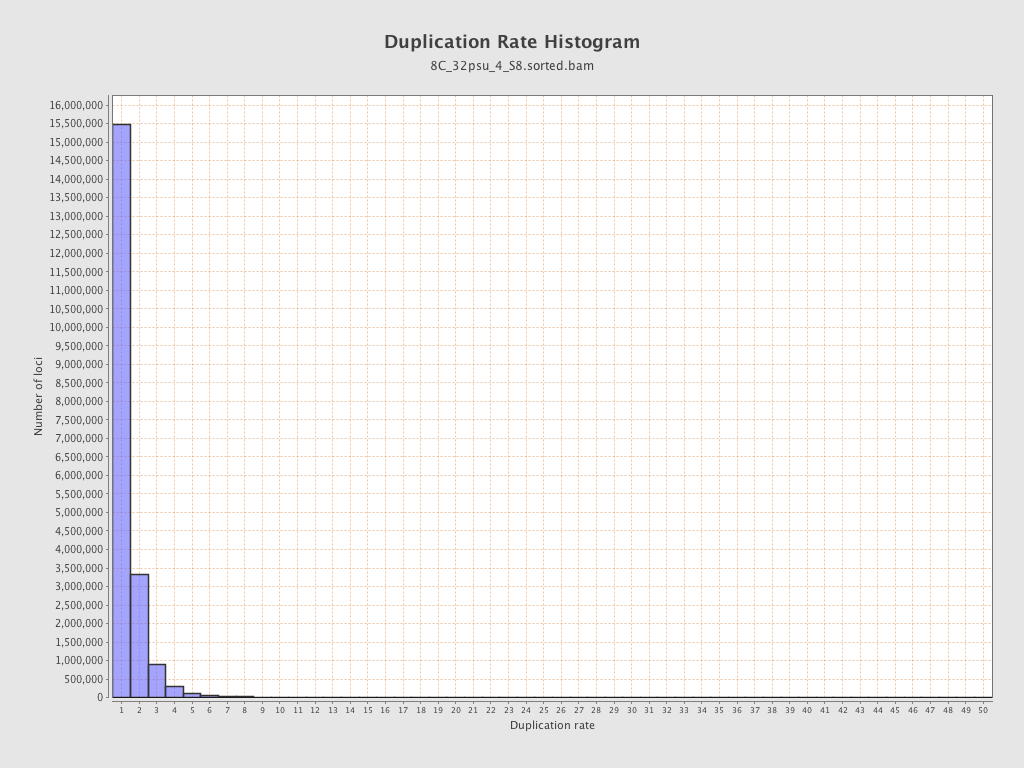
| Duplicated reads (estimated) | 8,303,004 / 29.03% |
| Duplication rate | 23.7% |
| Clipped reads | 0 / 0% |
ACGT Content
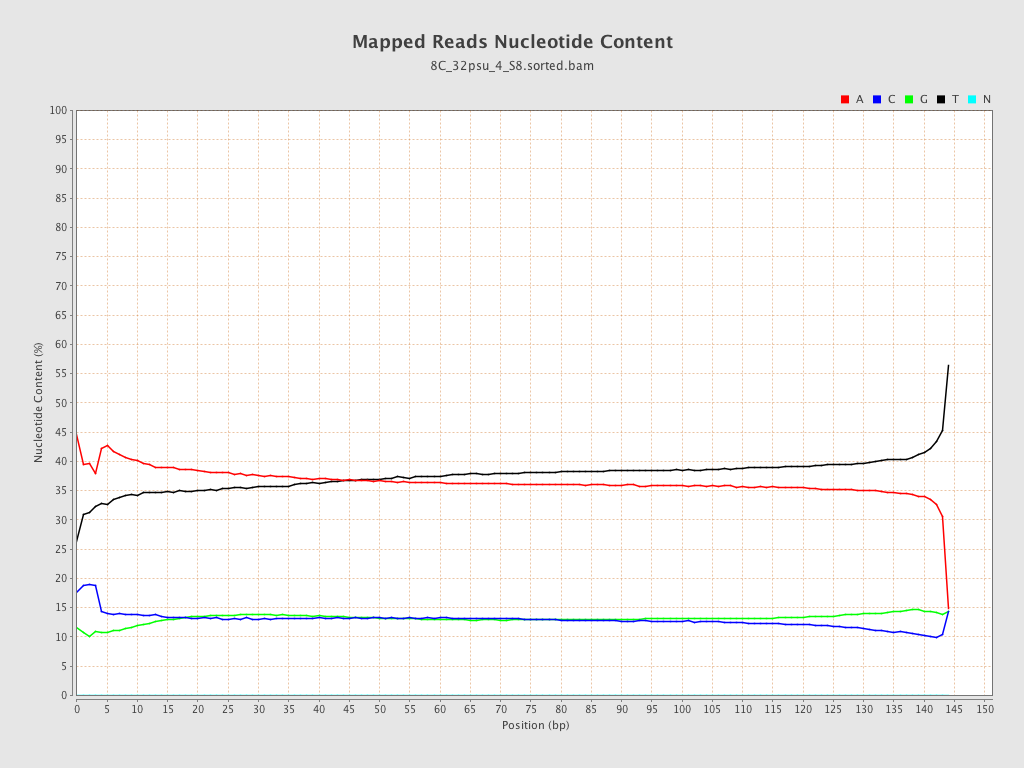
| Number/percentage of A's | 1,181,364,185 / 36.95% |
| Number/percentage of C's | 418,851,610 / 13.1% |
| Number/percentage of T's | 1,180,272,409 / 36.92% |
| Number/percentage of G's | 416,383,096 / 13.02% |
| Number/percentage of N's | 21,659 / 0% |
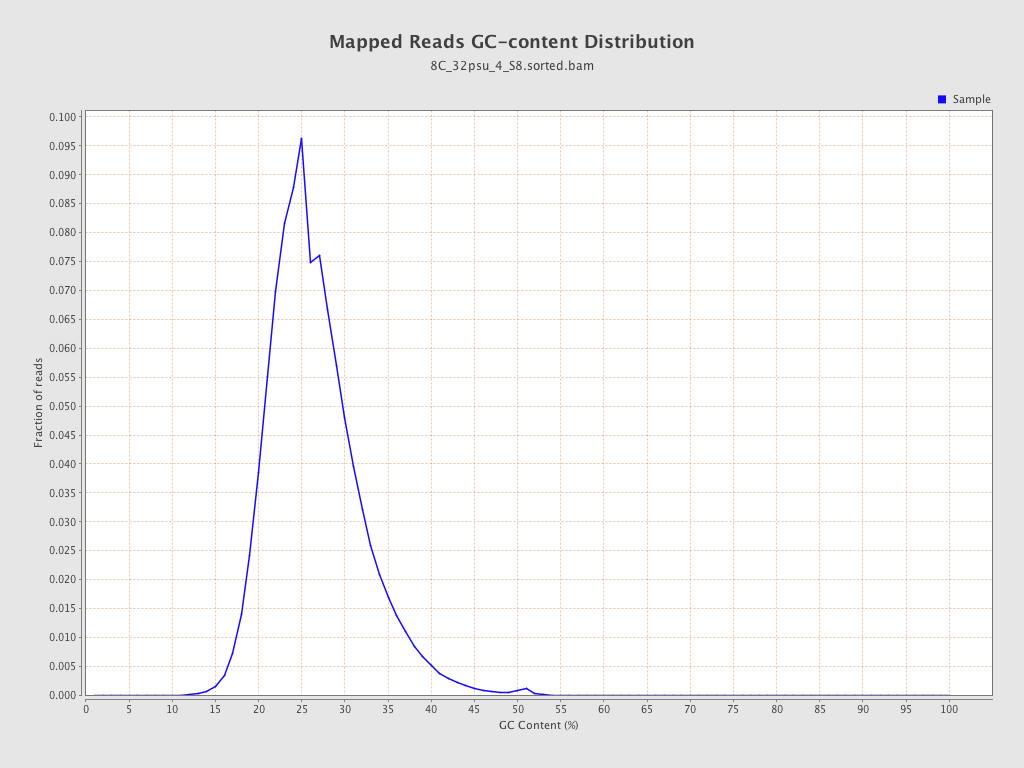
| GC Percentage | 26.13% |
Coverage
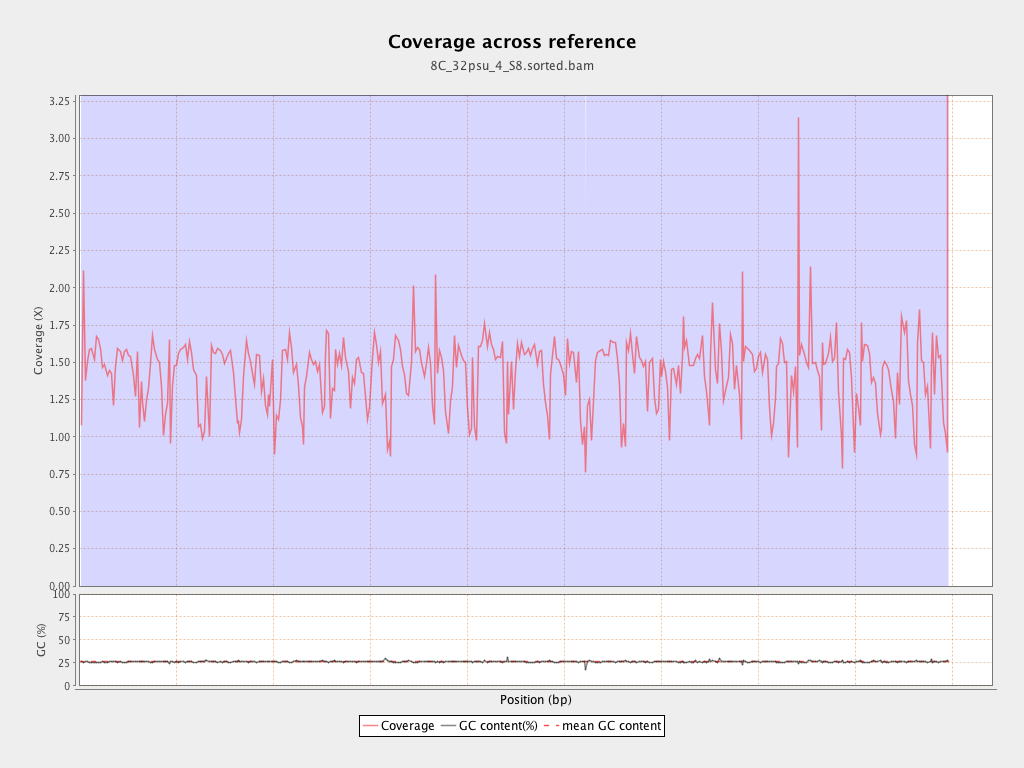
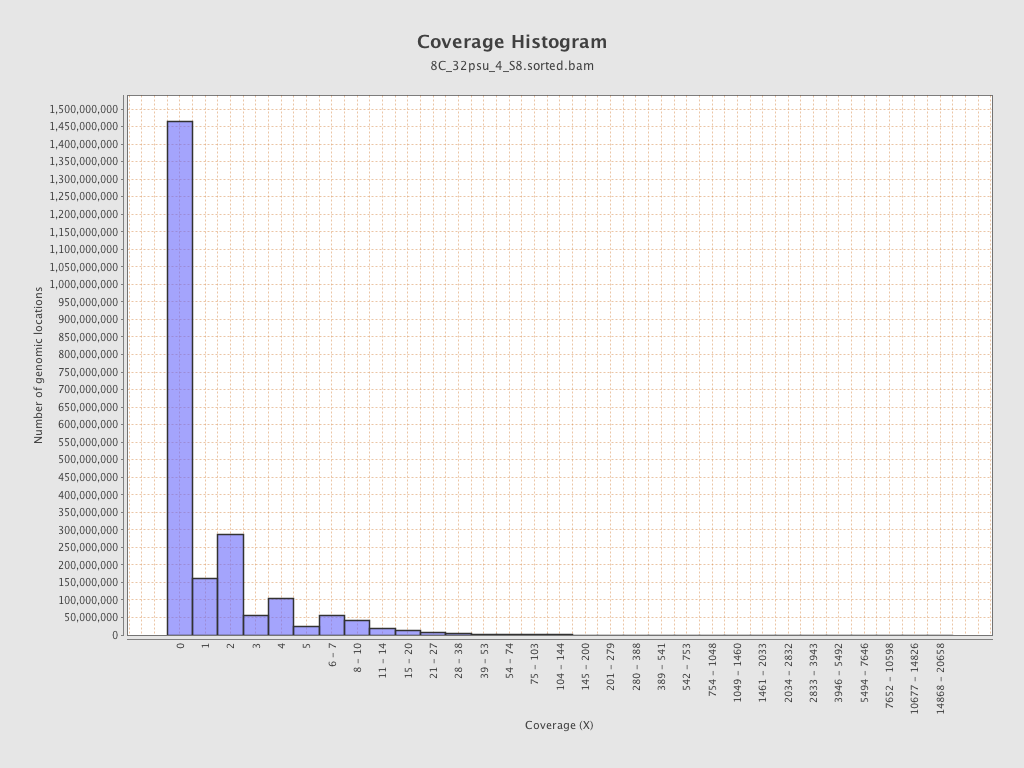
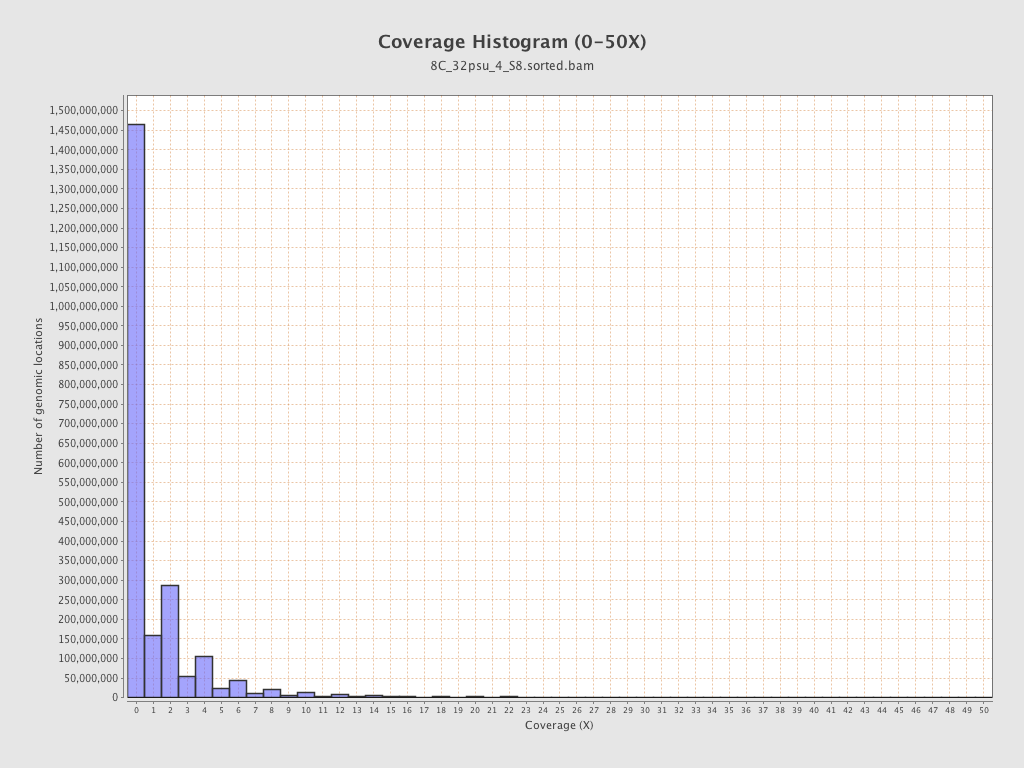
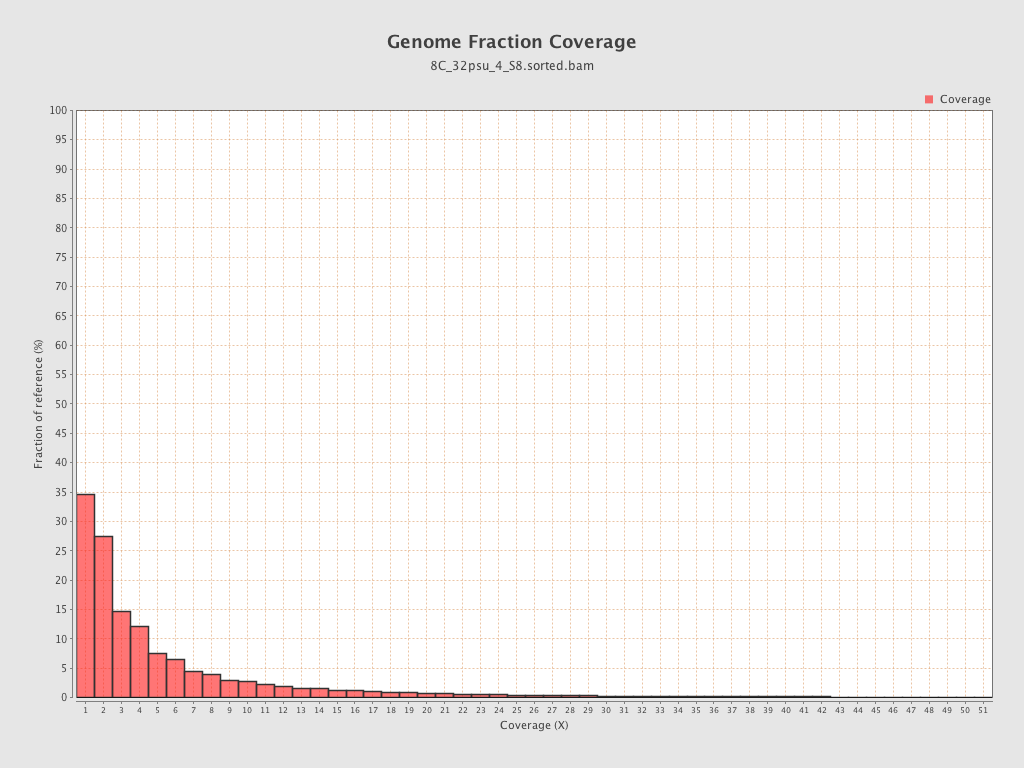
| Mean | 1.4312 |
| Standard Deviation | 11.4631 |
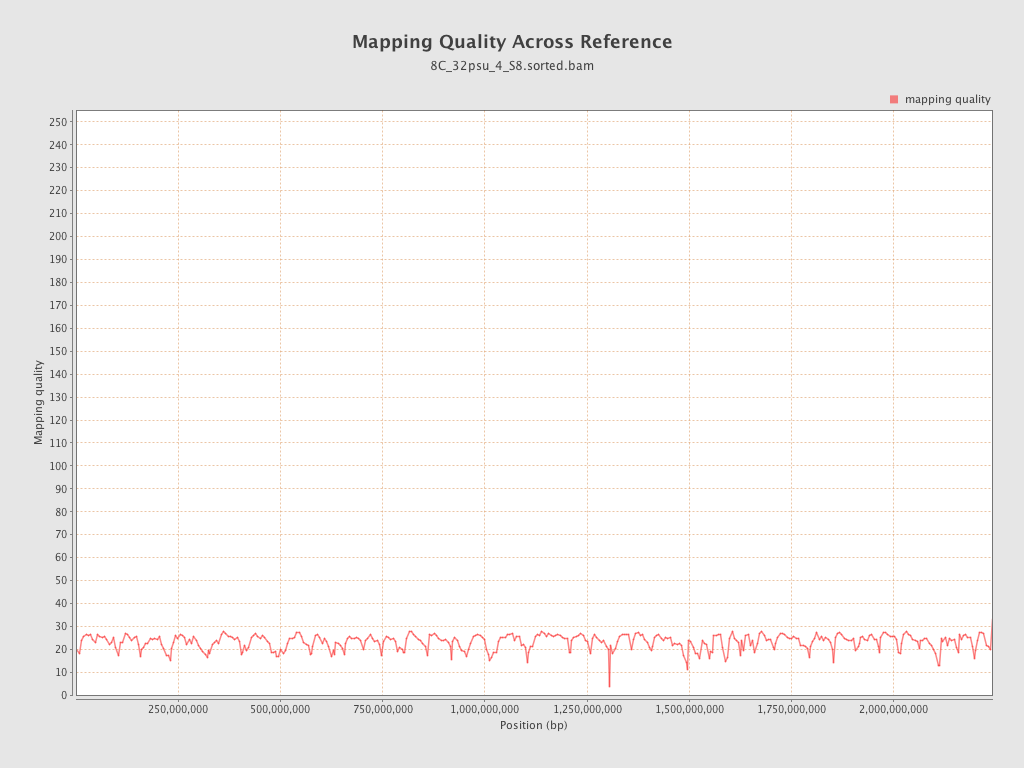
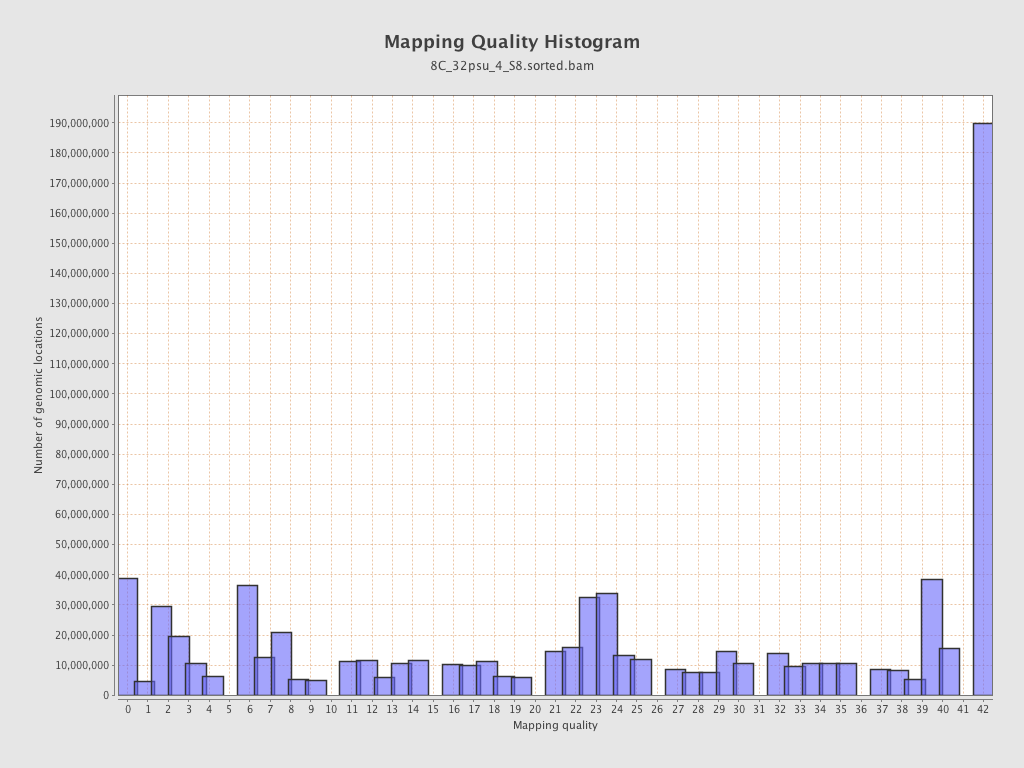
Mapping Quality
| Mean Mapping Quality | 23.08 |
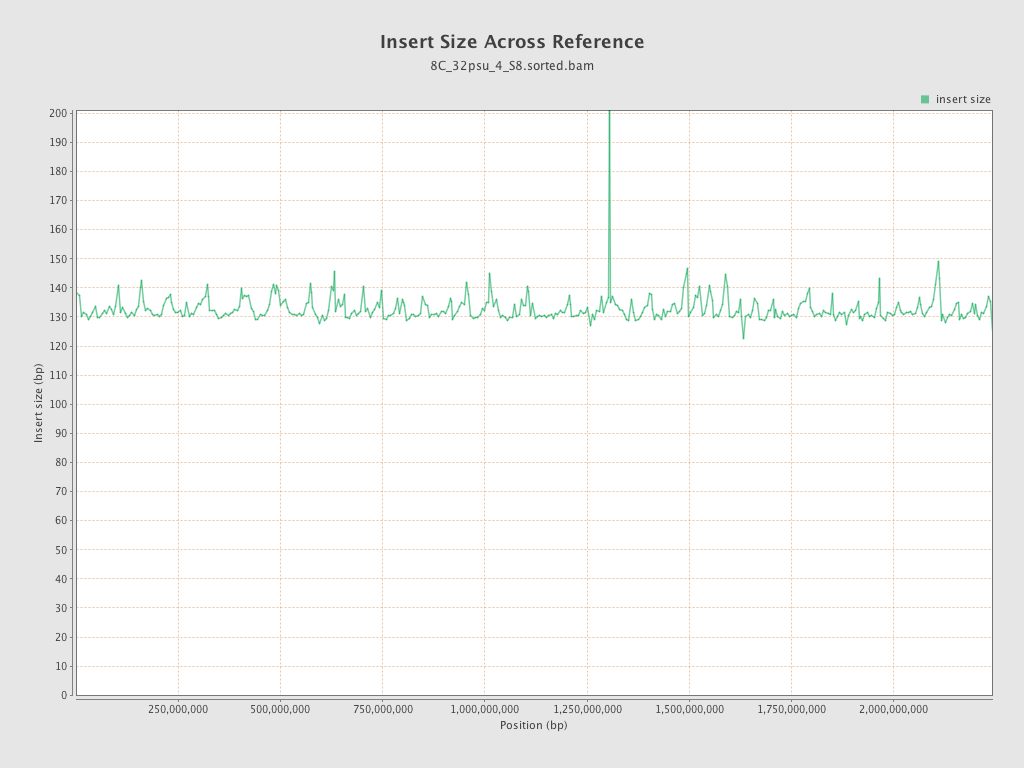
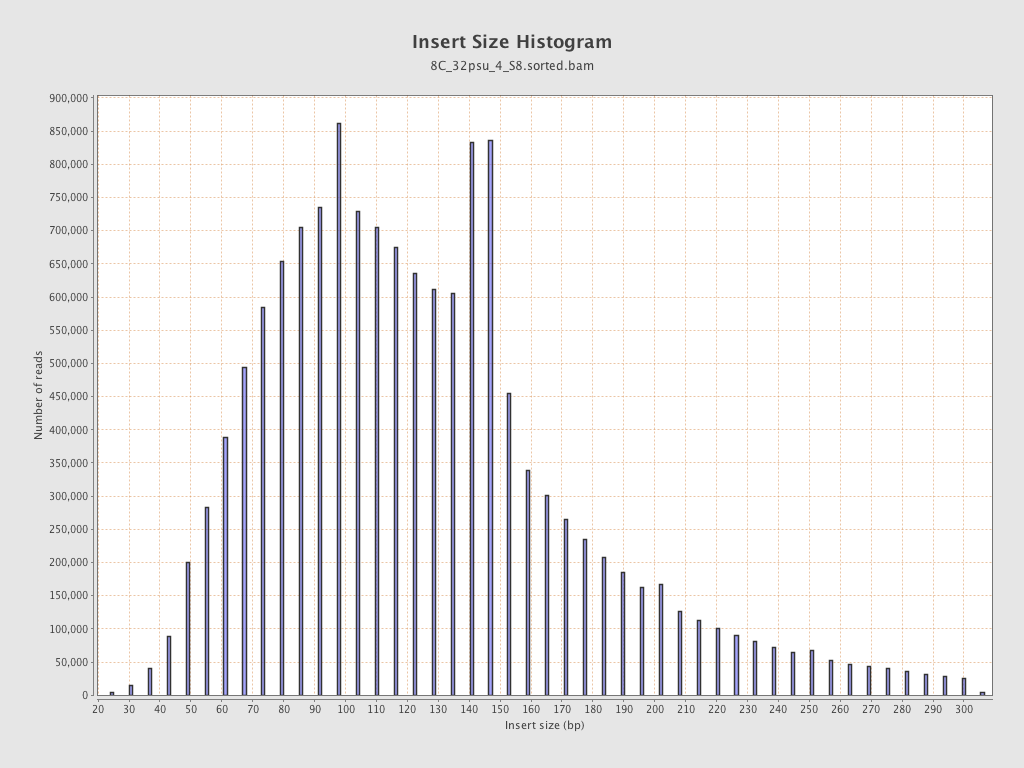
Insert size
| Mean | 132.72 |
| Standard Deviation | 59.51 |
| P25/Median/P75 | 93 / 122 / 153 |
Mismatches and indels
| General error rate | 21.92% |
| Mismatches | 674,659,390 |
| Insertions | 8,706,112 |
| Mapped reads with at least one insertion | 21.51% |
| Deletions | 5,735,343 |
| Mapped reads with at least one deletion | 17.05% |
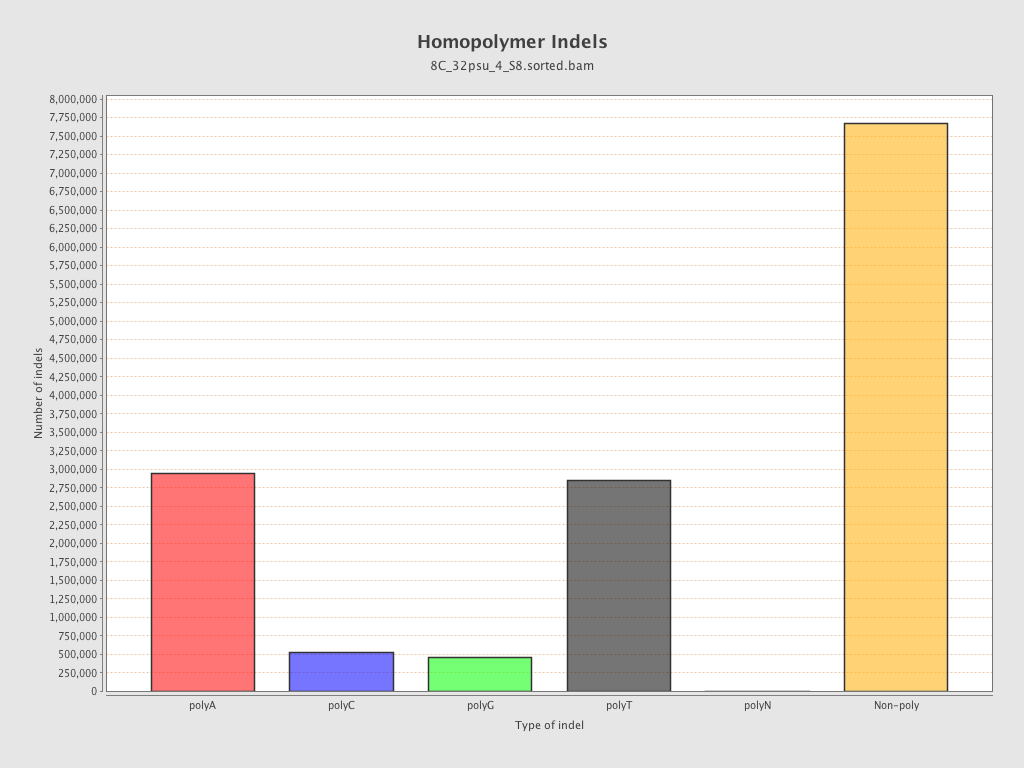
| Homopolymer indels | 46.9% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 237397167 | 1.4927 | 10.5173 |
| NC_027301.1 | 72943711 | 98686787 | 1.3529 | 5.4854 |
| NC_027302.1 | 92503428 | 128369659 | 1.3877 | 4.7653 |
| NC_027303.1 | 82398023 | 120406885 | 1.4613 | 9.0387 |
| NC_027304.1 | 80503876 | 109806937 | 1.364 | 7.218 |
| NC_027305.1 | 87043187 | 118006829 | 1.3557 | 11.2949 |
| NC_027306.1 | 58785265 | 81907553 | 1.3933 | 6.6818 |
| NC_027307.1 | 26434011 | 37941910 | 1.4353 | 10.7422 |
| NC_027308.1 | 141712163 | 197871967 | 1.3963 | 6.3703 |
| NC_027309.1 | 116138521 | 174228607 | 1.5002 | 22.4126 |
| NC_027310.1 | 93888508 | 130283632 | 1.3876 | 6.1995 |
| NC_027311.1 | 91880962 | 134548936 | 1.4644 | 11.058 |
| NC_027312.1 | 107758822 | 157632631 | 1.4628 | 6.4116 |
| NC_027313.1 | 93901823 | 133750011 | 1.4244 | 9.8233 |
| NC_027314.1 | 103963436 | 145106608 | 1.3957 | 4.586 |
| NC_027315.1 | 87796322 | 127772873 | 1.4553 | 7.0123 |
| NC_027316.1 | 57682537 | 79443062 | 1.3772 | 7.0426 |
| NC_027317.1 | 70695409 | 105510092 | 1.4925 | 8.528 |
| NC_027318.1 | 82978132 | 121307918 | 1.4619 | 24.7042 |
| NC_027319.1 | 86795997 | 124779121 | 1.4376 | 6.8452 |
| NC_027320.1 | 58021487 | 80530372 | 1.3879 | 4.9738 |
| NC_027321.1 | 63420196 | 97855077 | 1.543 | 23.7102 |
| NC_027322.1 | 49854004 | 73517243 | 1.4747 | 6.7816 |
| NC_027323.1 | 48650976 | 65519509 | 1.3467 | 4.6974 |
| NC_027324.1 | 51481326 | 72643740 | 1.4111 | 5.5752 |
| NC_027325.1 | 47900953 | 64582059 | 1.3482 | 6.4686 |
| NC_027326.1 | 43943985 | 62658384 | 1.4259 | 7.1902 |
| NC_027327.1 | 39600944 | 56553296 | 1.4281 | 8.1133 |
| NC_027328.1 | 42488238 | 56340485 | 1.326 | 11.2793 |
| NC_001960.1 | 16665 | 11247438 | 674.9138 | 1,129.8675 |