Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 25,557,502 |
| Mapped reads | 25,557,502 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 25,557,502 / 100% |
| Mapped reads, first in pair | 12,778,751 / 50% |
| Mapped reads, second in pair | 12,778,751 / 50% |
| Mapped reads, both in pair | 25,557,502 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 117.79 |
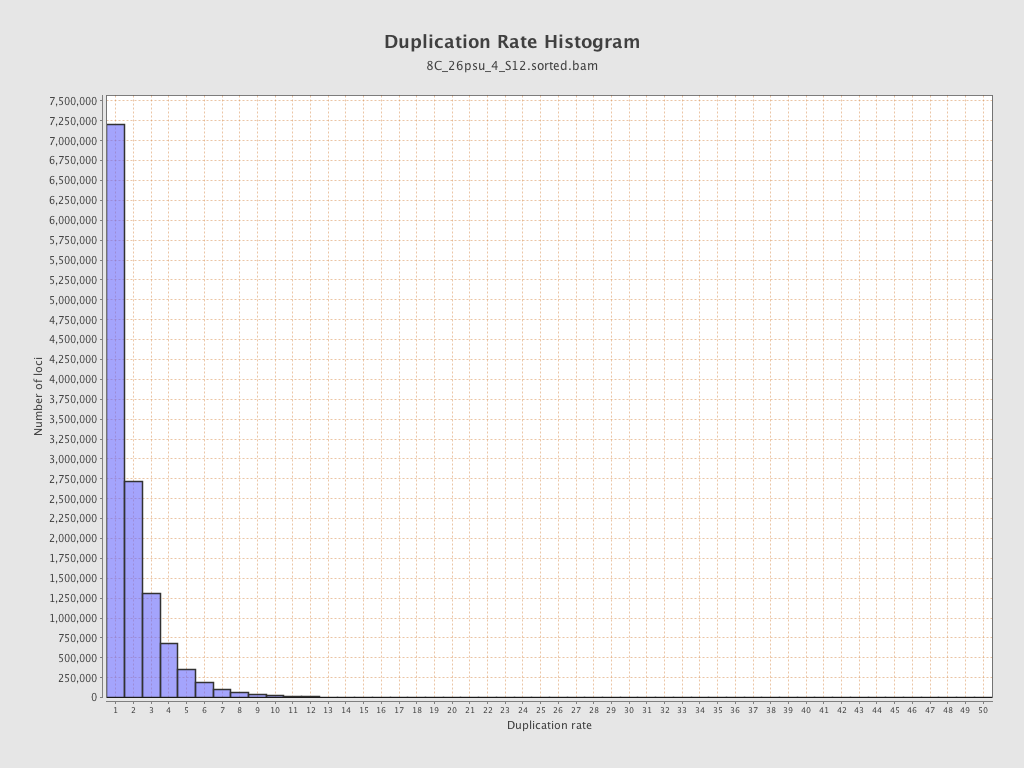
| Duplicated reads (estimated) | 12,808,795 / 50.12% |
| Duplication rate | 43.44% |
| Clipped reads | 0 / 0% |
ACGT Content
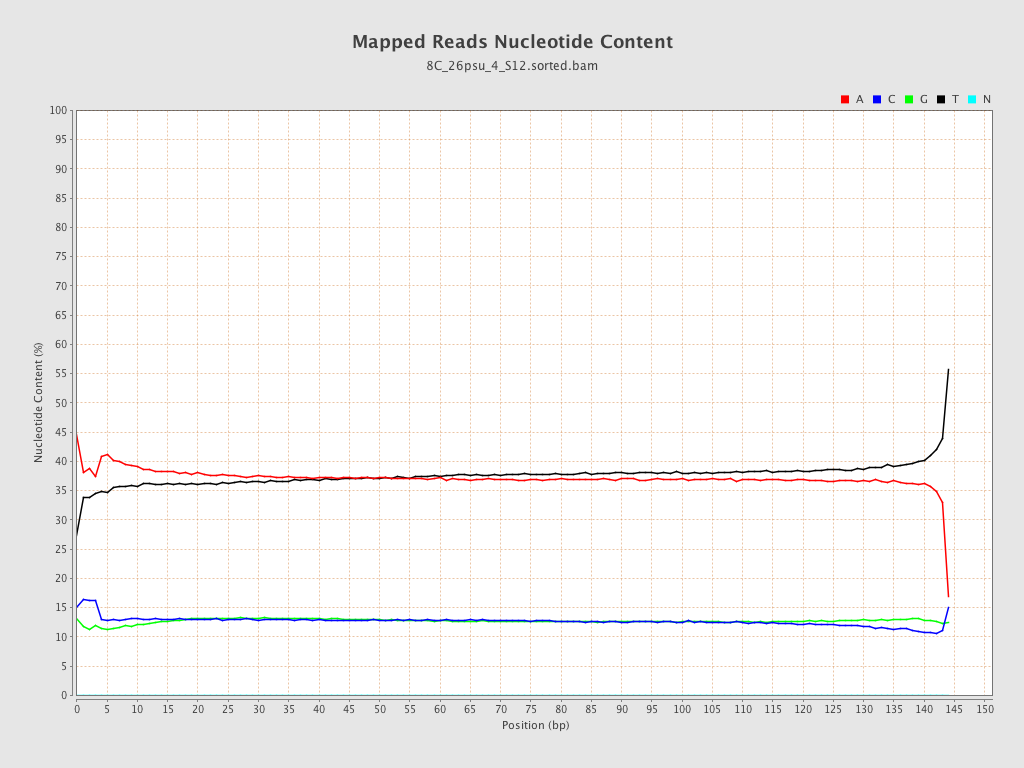
| Number/percentage of A's | 1,116,622,296 / 37.31% |
| Number/percentage of C's | 381,649,756 / 12.75% |
| Number/percentage of T's | 1,114,662,567 / 37.25% |
| Number/percentage of G's | 379,754,008 / 12.69% |
| Number/percentage of N's | 21,014 / 0% |
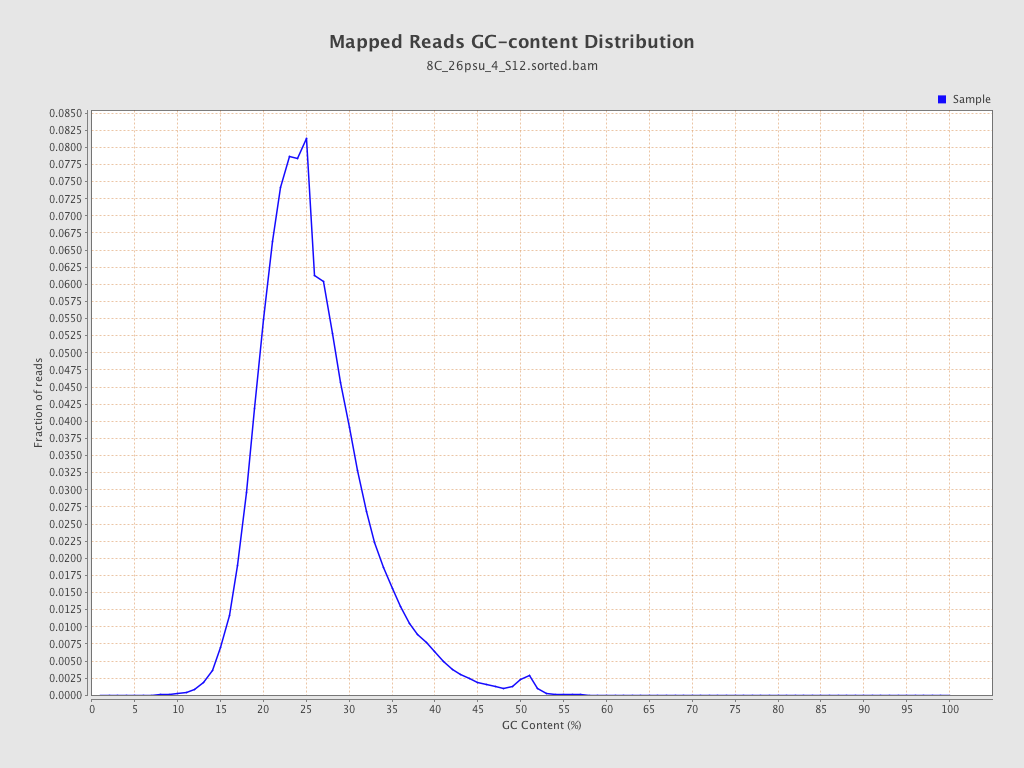
| GC Percentage | 25.44% |
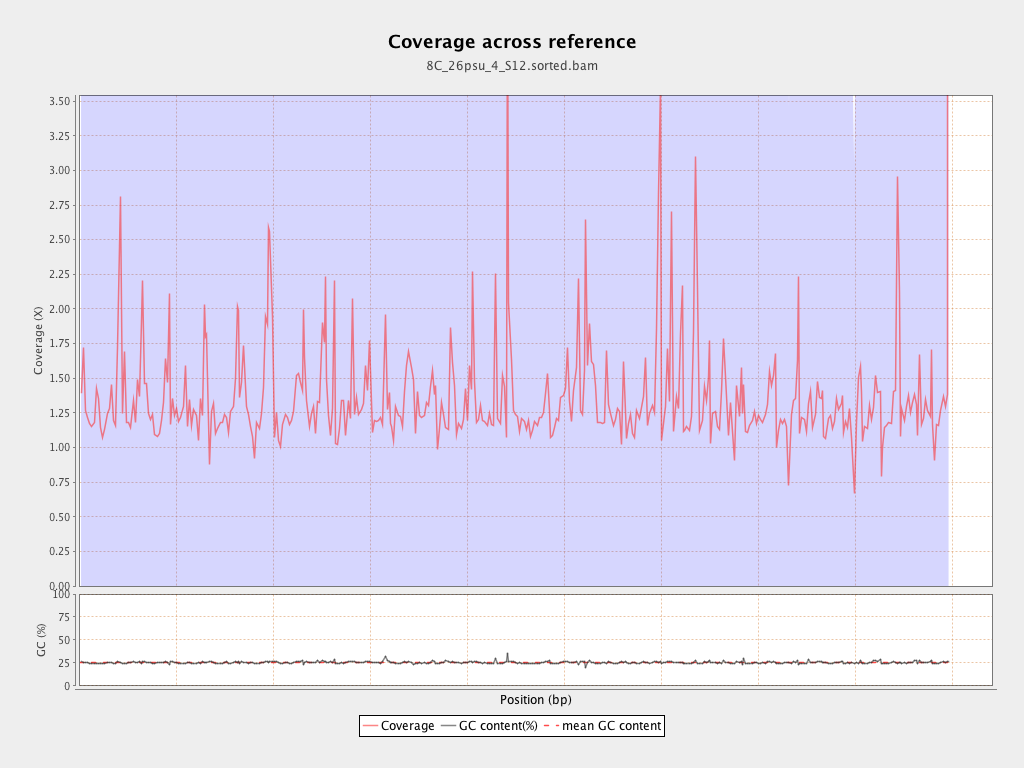
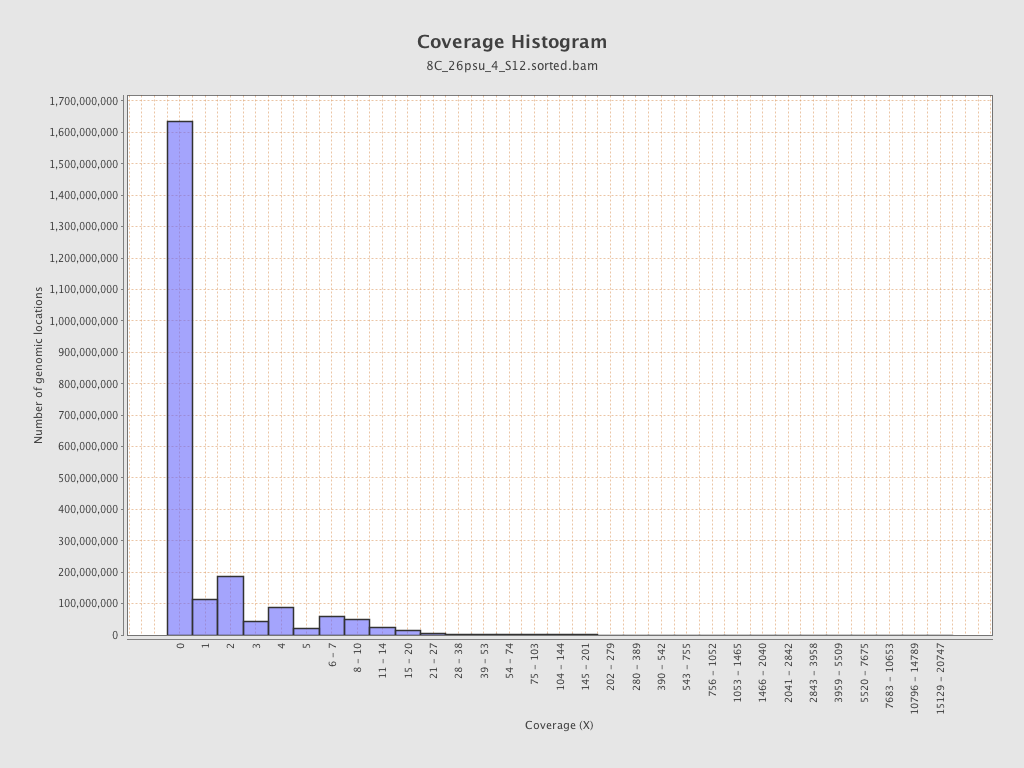
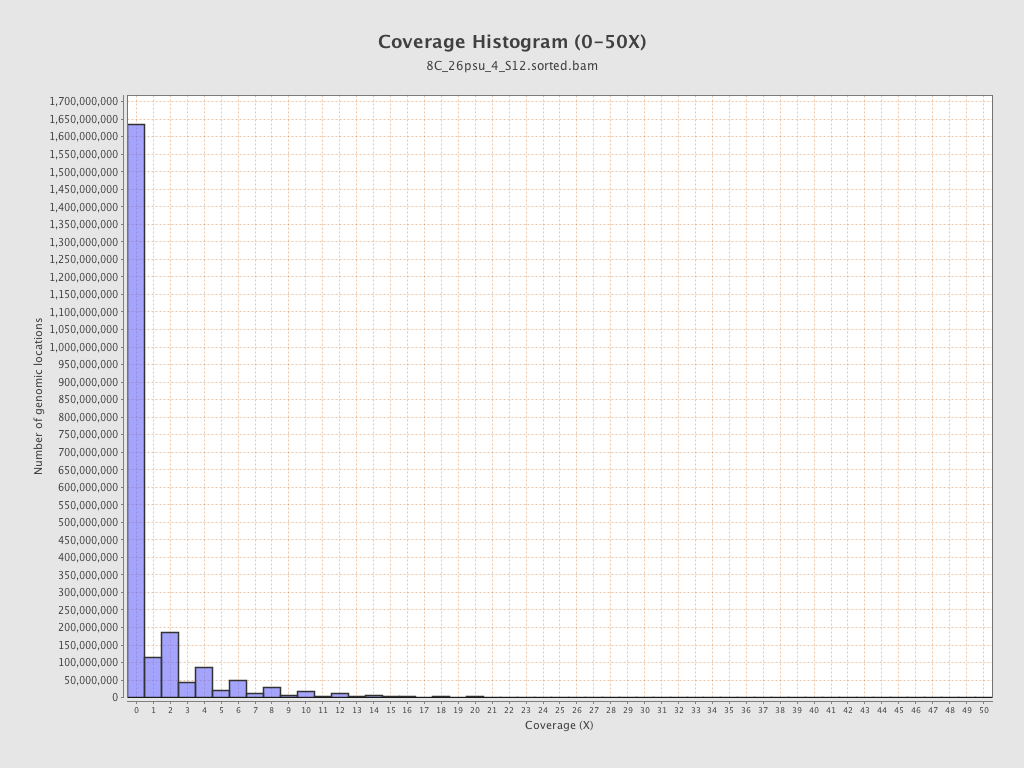
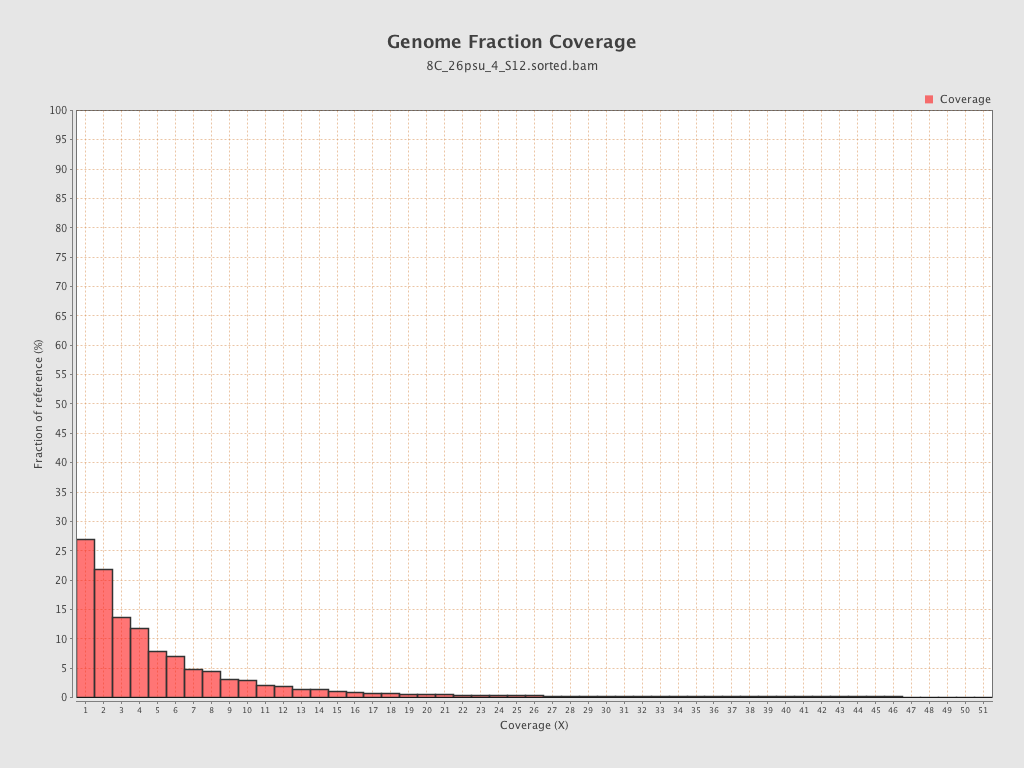
Coverage
| Mean | 1.34 |
| Standard Deviation | 13.3278 |
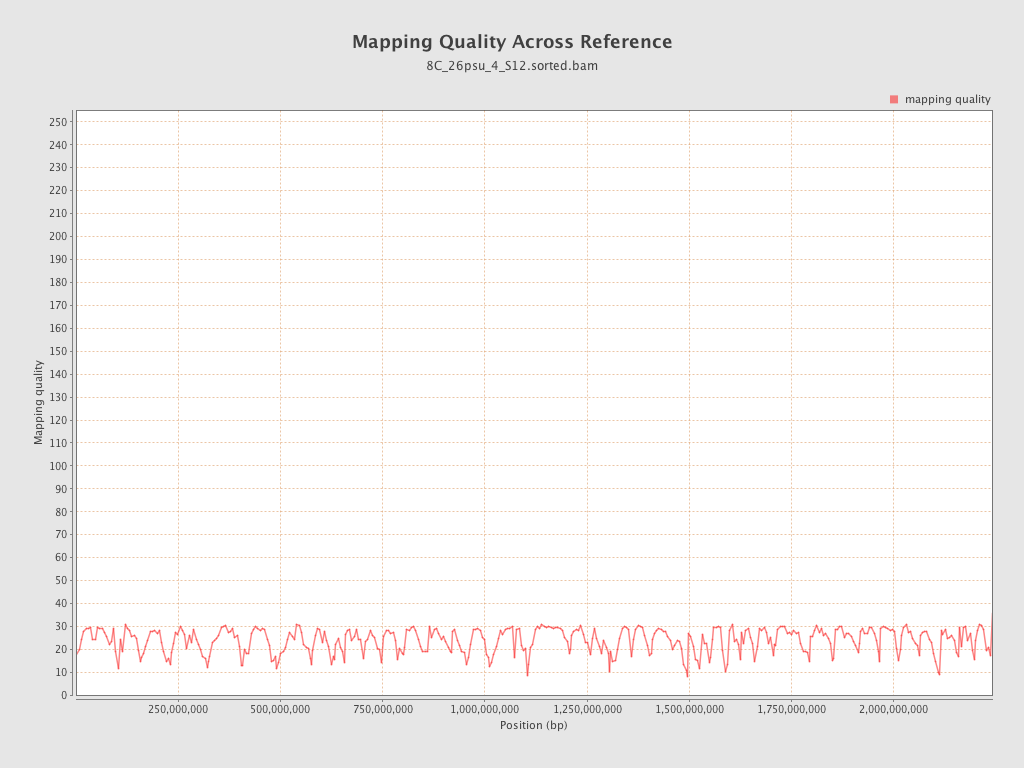
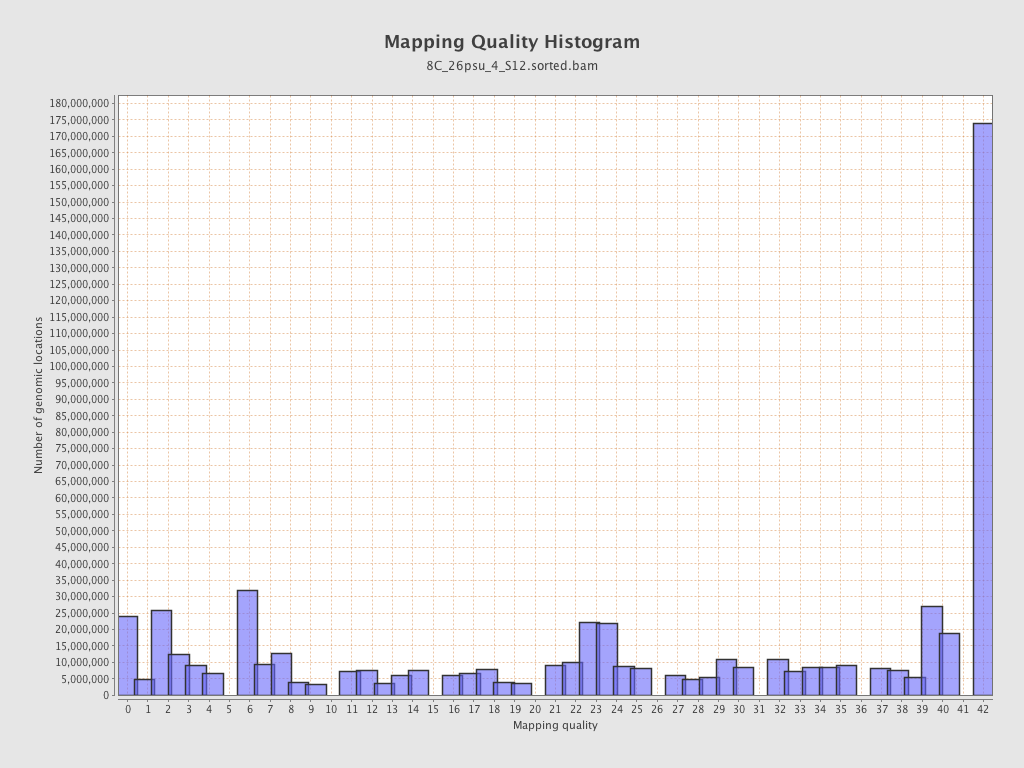
Mapping Quality
| Mean Mapping Quality | 23.74 |
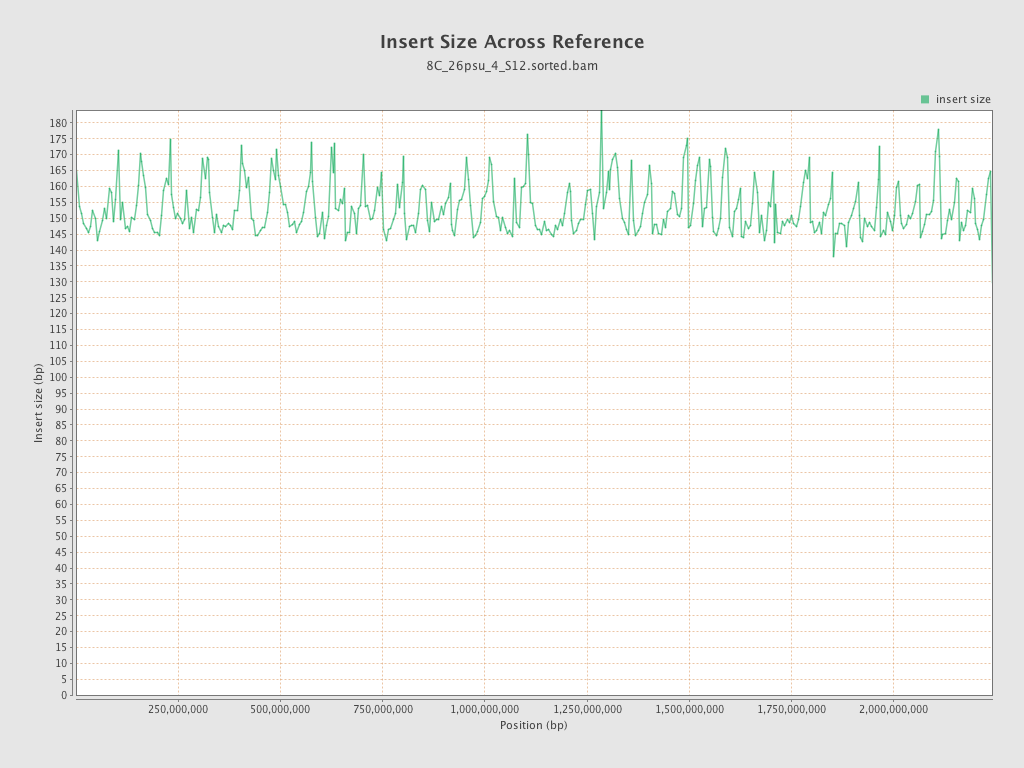
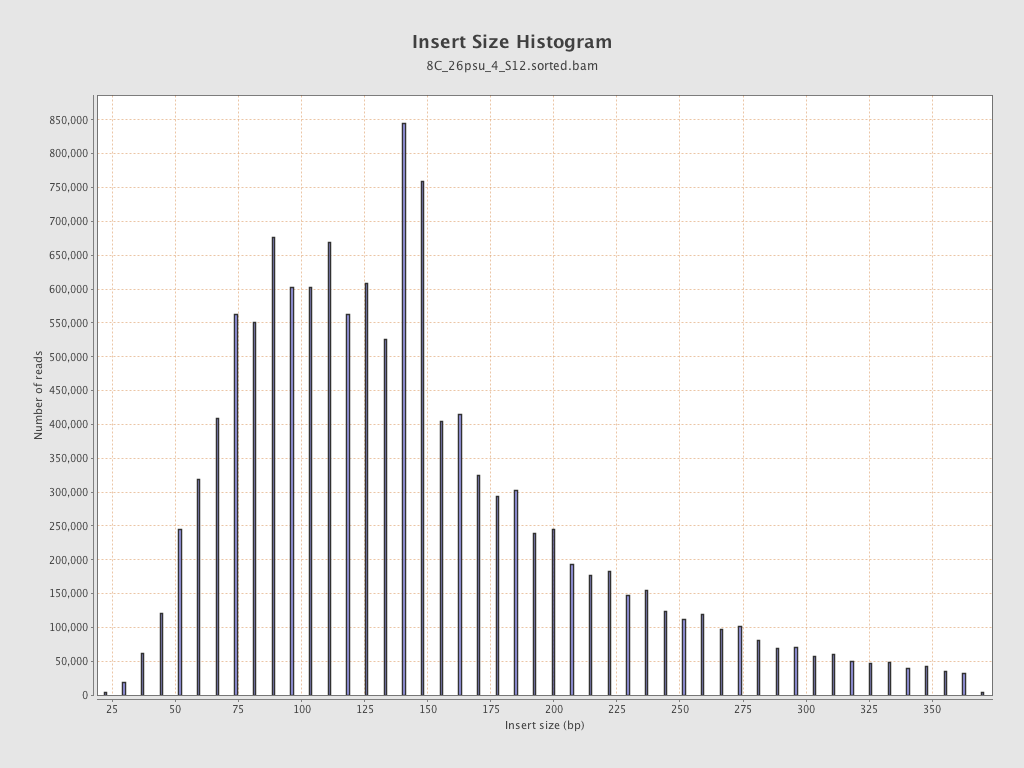
Insert size
| Mean | 154.65 |
| Standard Deviation | 80.35 |
| P25/Median/P75 | 99 / 139 / 185 |
Mismatches and indels
| General error rate | 20.47% |
| Mismatches | 595,597,068 |
| Insertions | 6,060,286 |
| Mapped reads with at least one insertion | 16.95% |
| Deletions | 5,322,998 |
| Mapped reads with at least one deletion | 17.4% |
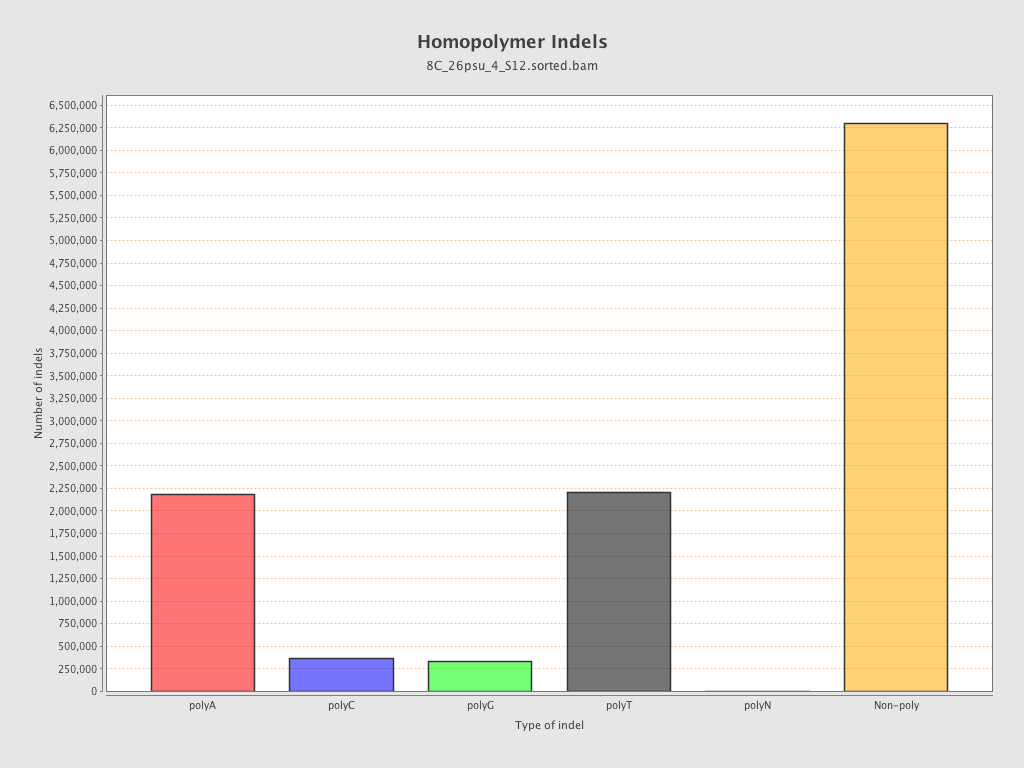
| Homopolymer indels | 44.7% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 216553533 | 1.3616 | 14.3744 |
| NC_027301.1 | 72943711 | 99273706 | 1.361 | 9.3189 |
| NC_027302.1 | 92503428 | 120599983 | 1.3037 | 9.7636 |
| NC_027303.1 | 82398023 | 105882237 | 1.285 | 9.7184 |
| NC_027304.1 | 80503876 | 111124091 | 1.3804 | 10.1179 |
| NC_027305.1 | 87043187 | 119680826 | 1.375 | 14.8193 |
| NC_027306.1 | 58785265 | 84603391 | 1.4392 | 11.234 |
| NC_027307.1 | 26434011 | 37673937 | 1.4252 | 20.0276 |
| NC_027308.1 | 141712163 | 189126784 | 1.3346 | 11.7635 |
| NC_027309.1 | 116138521 | 155092578 | 1.3354 | 16.6515 |
| NC_027310.1 | 93888508 | 122022643 | 1.2997 | 9.6761 |
| NC_027311.1 | 91880962 | 122529985 | 1.3336 | 17.6969 |
| NC_027312.1 | 107758822 | 138519715 | 1.2855 | 8.8749 |
| NC_027313.1 | 93901823 | 129527868 | 1.3794 | 26.8098 |
| NC_027314.1 | 103963436 | 142105467 | 1.3669 | 10.3301 |
| NC_027315.1 | 87796322 | 125121792 | 1.4251 | 12.4591 |
| NC_027316.1 | 57682537 | 88314169 | 1.531 | 13.9671 |
| NC_027317.1 | 70695409 | 106976751 | 1.5132 | 15.5648 |
| NC_027318.1 | 82978132 | 106343042 | 1.2816 | 10.06 |
| NC_027319.1 | 86795997 | 111843767 | 1.2886 | 9.649 |
| NC_027320.1 | 58021487 | 69434165 | 1.1967 | 5.9719 |
| NC_027321.1 | 63420196 | 81247497 | 1.2811 | 12.5819 |
| NC_027322.1 | 49854004 | 60268685 | 1.2089 | 6.1158 |
| NC_027323.1 | 48650976 | 57497267 | 1.1818 | 10.5784 |
| NC_027324.1 | 51481326 | 65556214 | 1.2734 | 9.8006 |
| NC_027325.1 | 47900953 | 70098881 | 1.4634 | 18.0172 |
| NC_027326.1 | 43943985 | 56551534 | 1.2869 | 5.25 |
| NC_027327.1 | 39600944 | 52113816 | 1.316 | 19.0601 |
| NC_027328.1 | 42488238 | 51668442 | 1.2161 | 6.9987 |
| NC_001960.1 | 16665 | 4594714 | 275.7104 | 421.5955 |