Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 38,997,124 |
| Mapped reads | 38,997,124 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 38,997,124 / 100% |
| Mapped reads, first in pair | 19,498,562 / 50% |
| Mapped reads, second in pair | 19,498,562 / 50% |
| Mapped reads, both in pair | 38,997,124 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 106.76 |
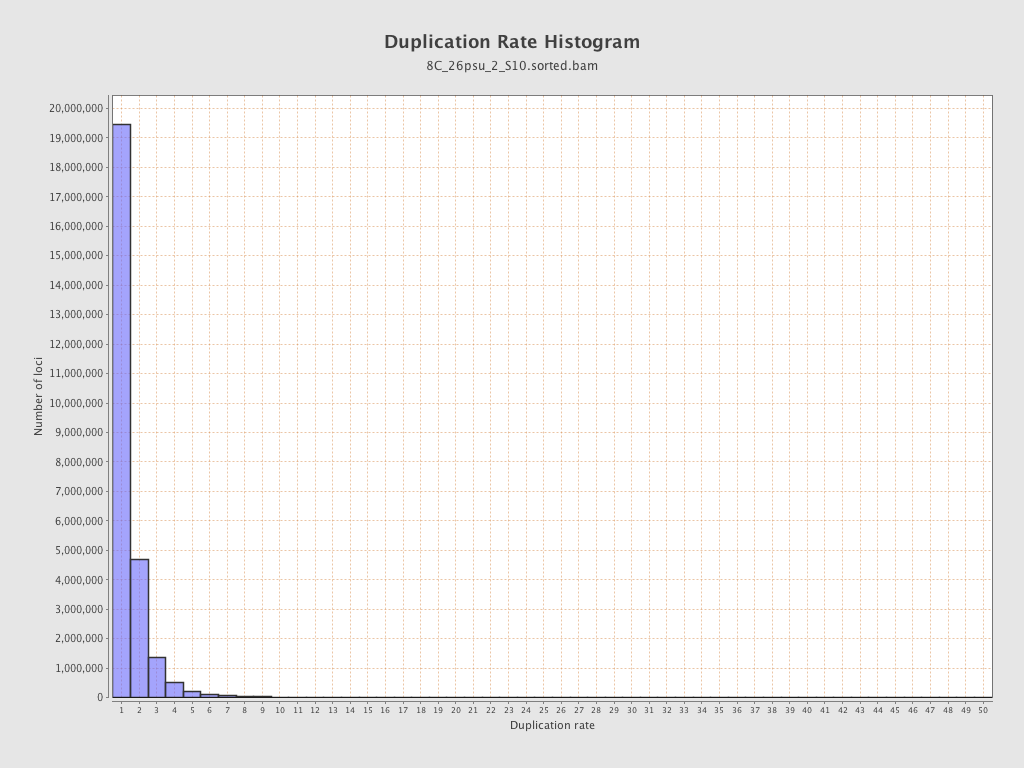
| Duplicated reads (estimated) | 12,481,258 / 32.01% |
| Duplication rate | 26.57% |
| Clipped reads | 0 / 0% |
ACGT Content
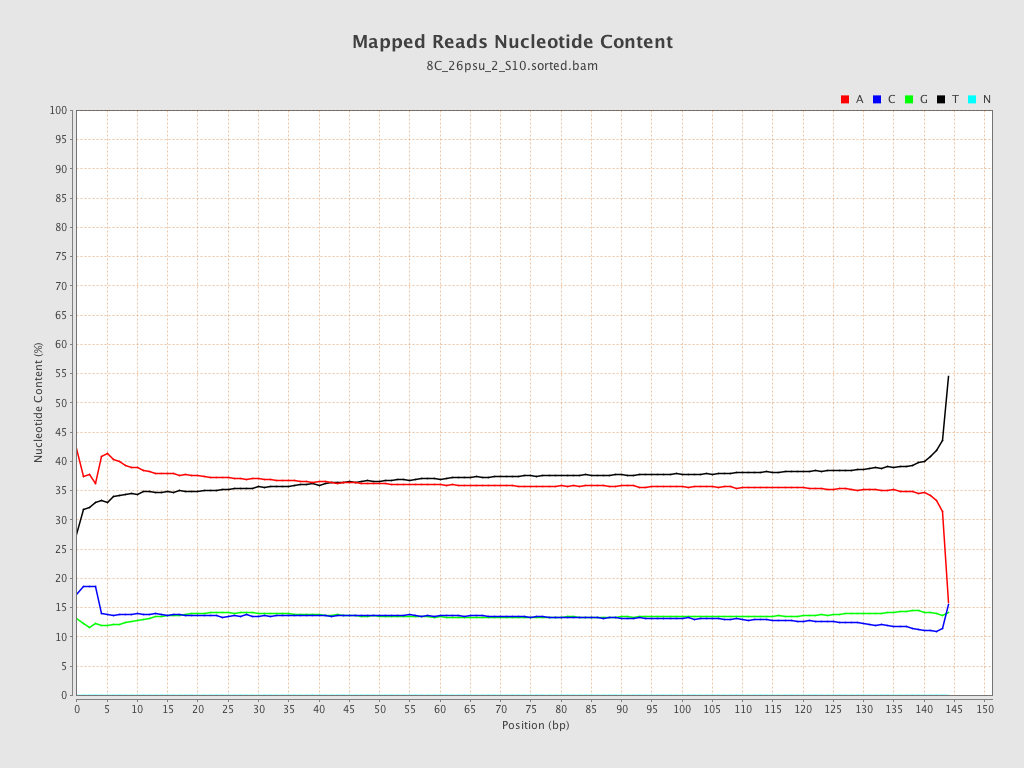
| Number/percentage of A's | 1,510,679,015 / 36.52% |
| Number/percentage of C's | 560,826,315 / 13.56% |
| Number/percentage of T's | 1,507,996,841 / 36.46% |
| Number/percentage of G's | 556,975,000 / 13.46% |
| Number/percentage of N's | 26,257 / 0% |
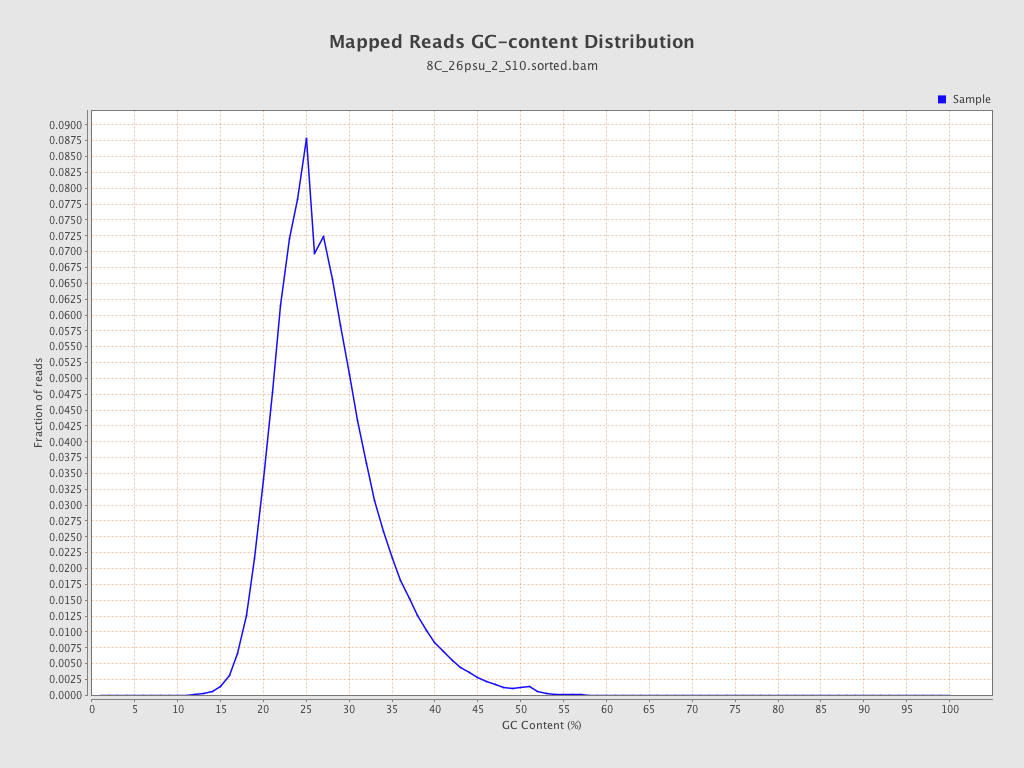
| GC Percentage | 27.02% |
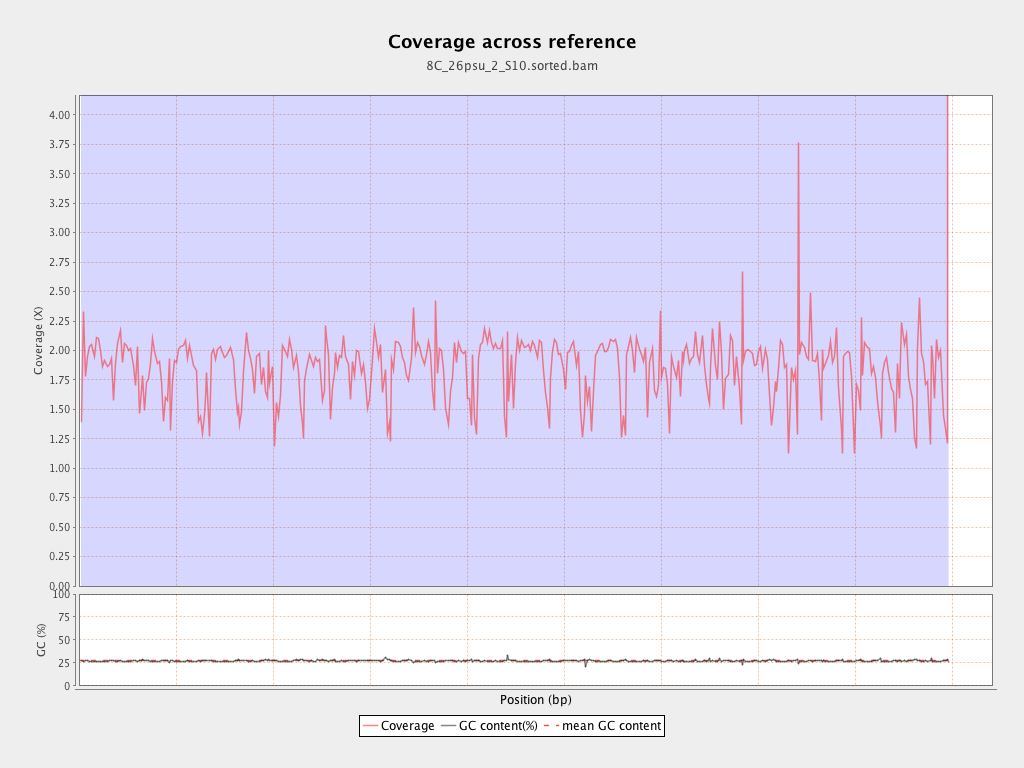
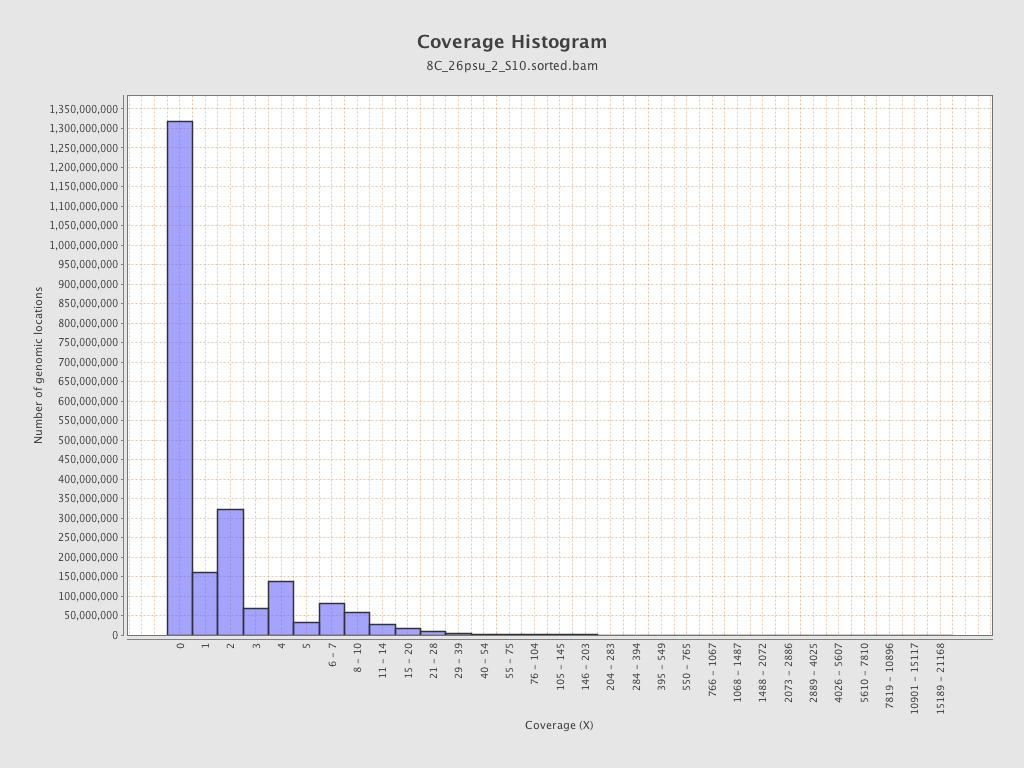
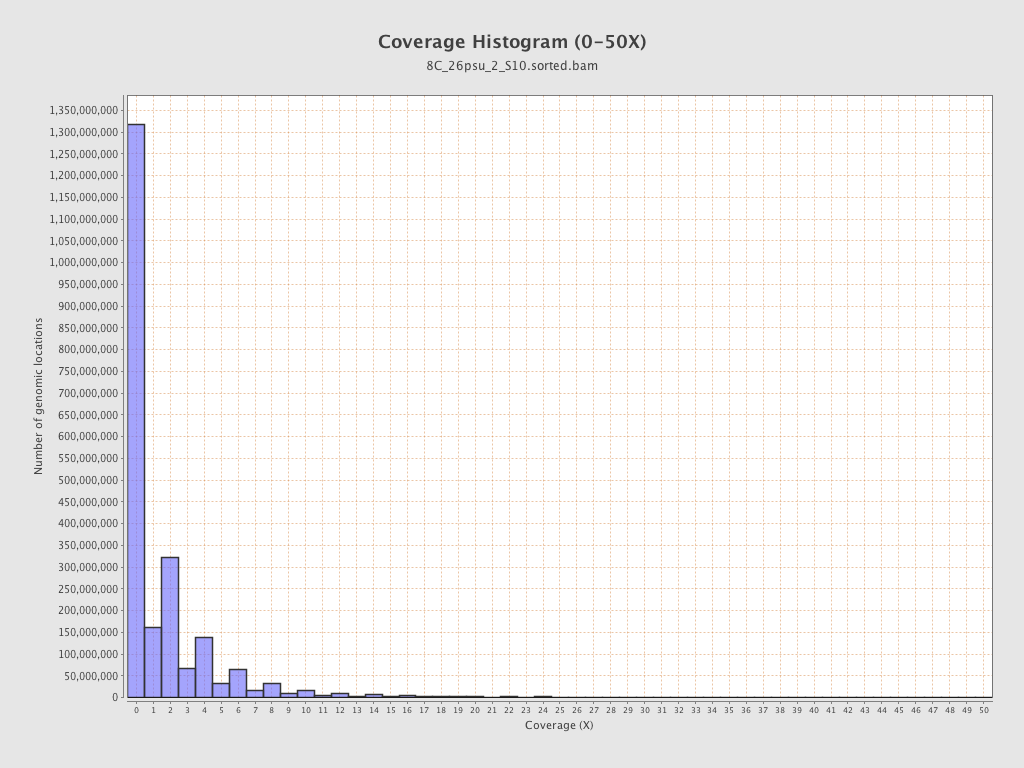
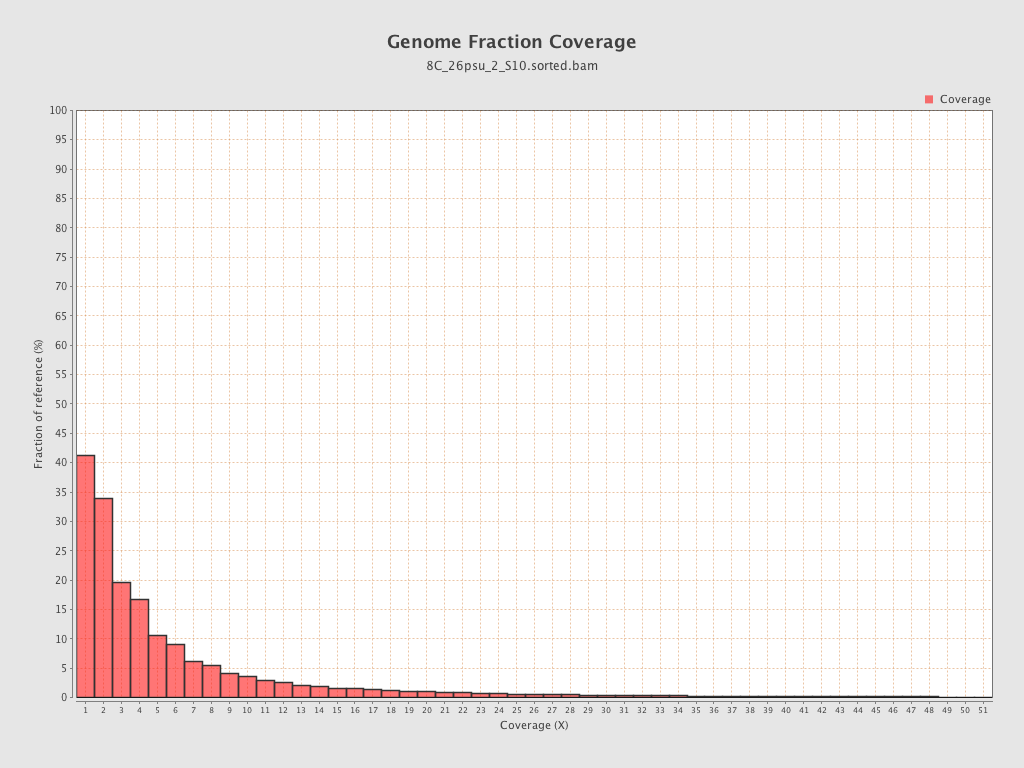
Coverage
| Mean | 1.8514 |
| Standard Deviation | 12.4513 |
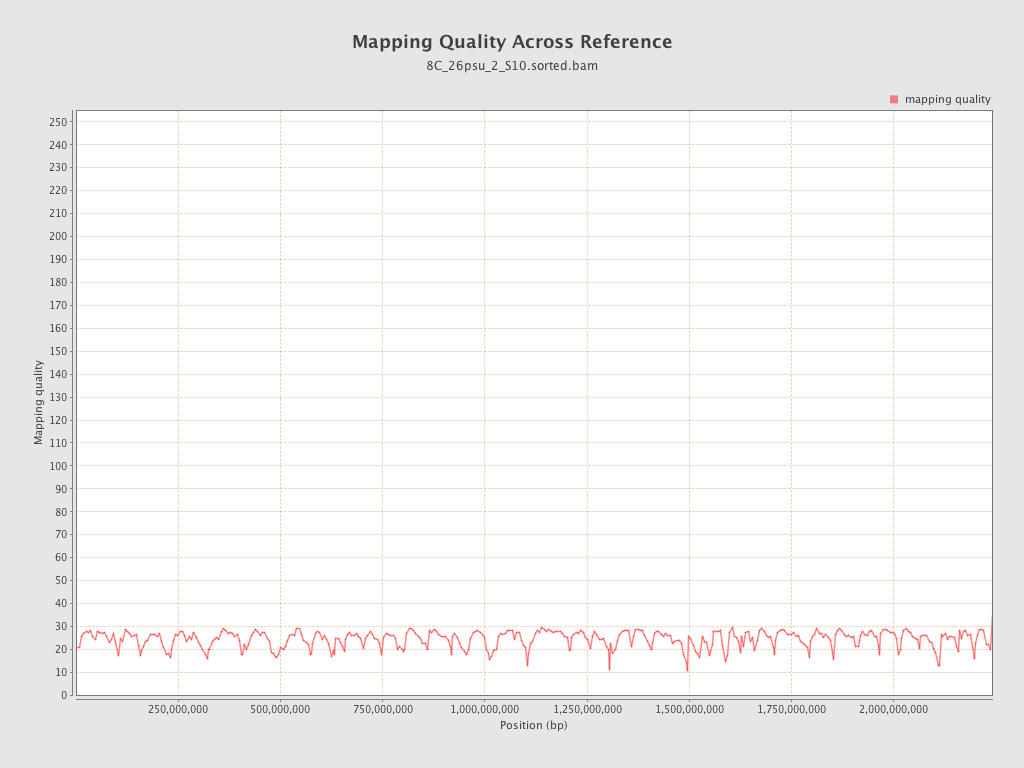
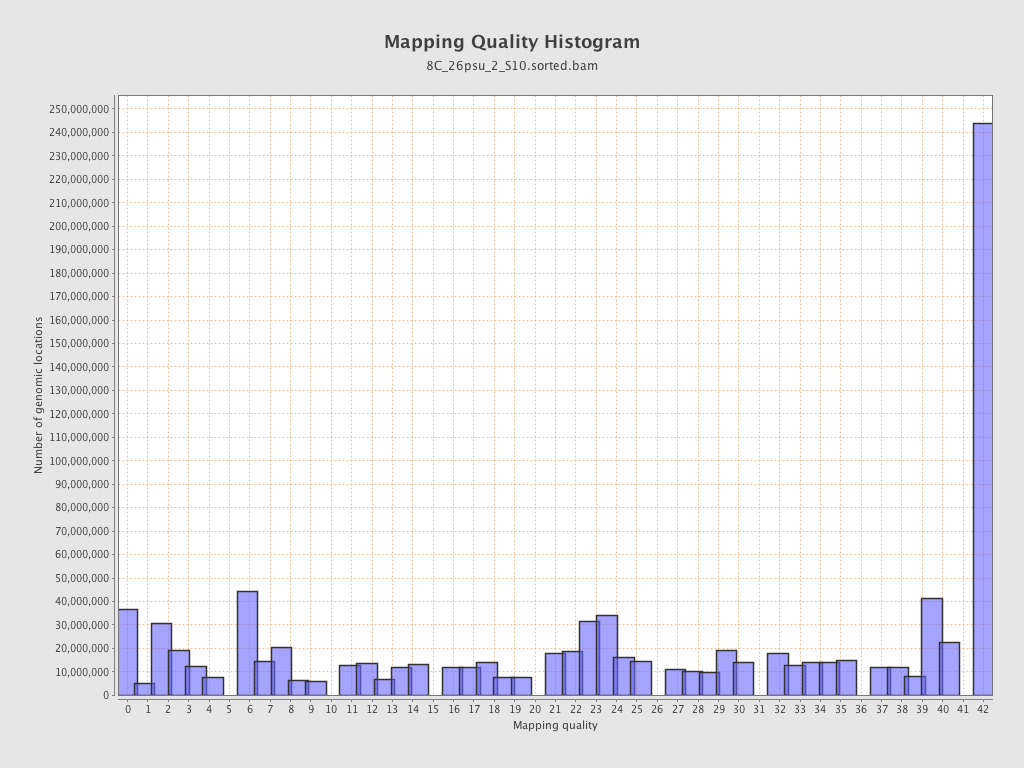
Mapping Quality
| Mean Mapping Quality | 24.27 |
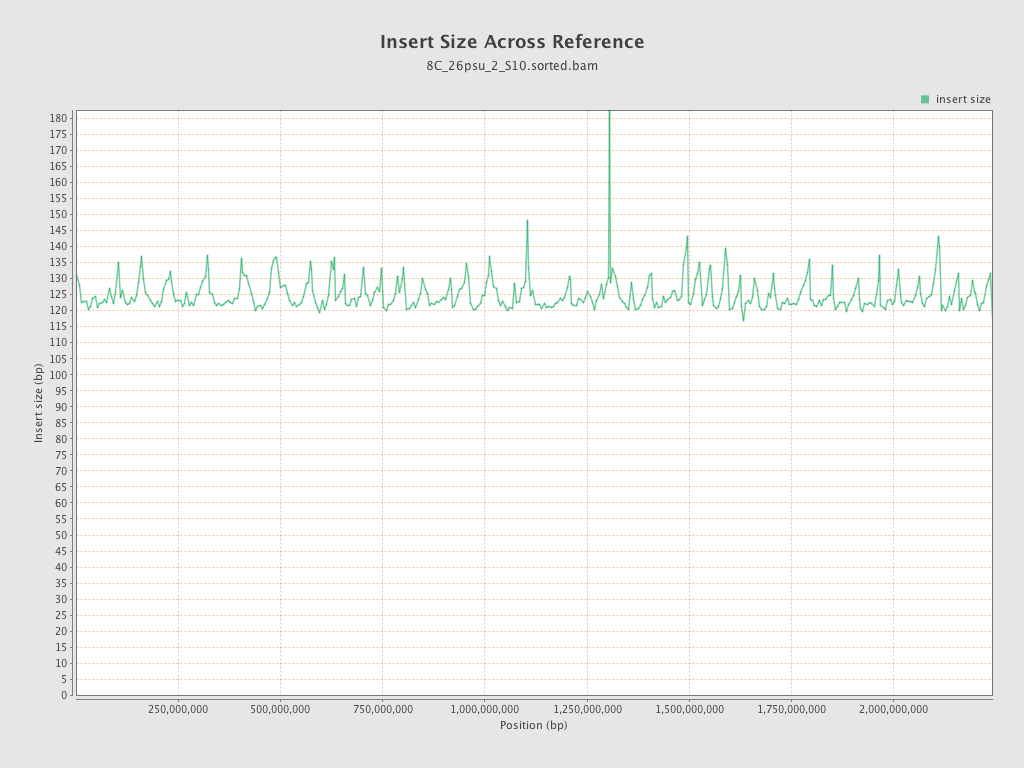
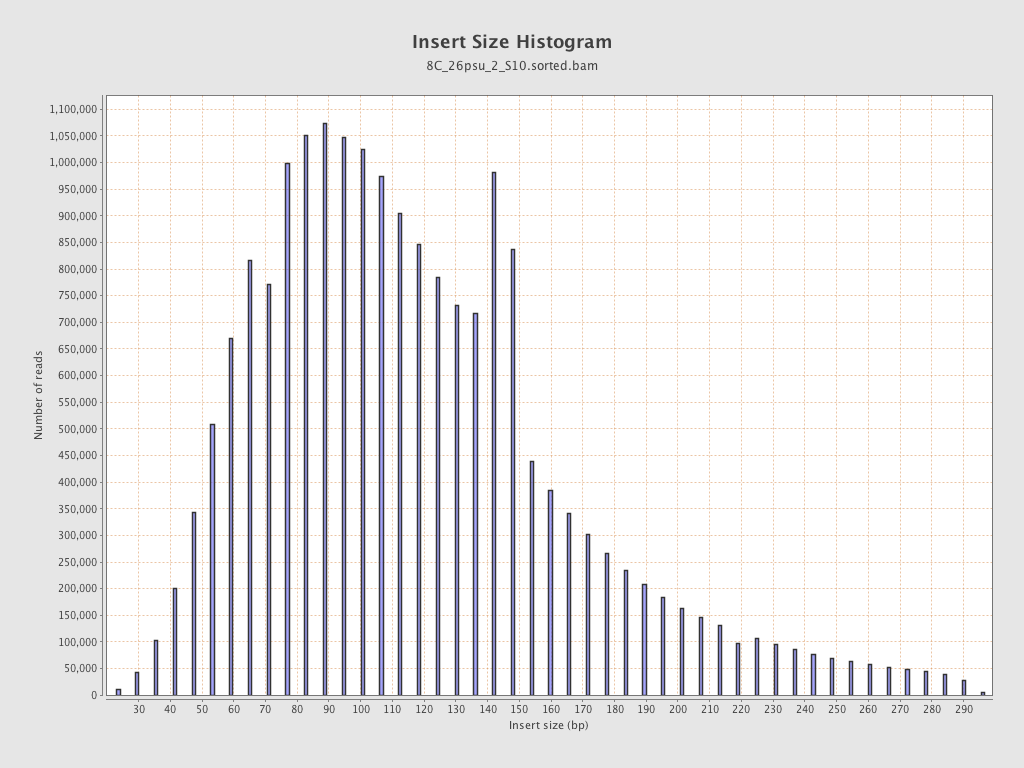
Insert size
| Mean | 124.88 |
| Standard Deviation | 59.4 |
| P25/Median/P75 | 85 / 113 / 148 |
Mismatches and indels
| General error rate | 20.87% |
| Mismatches | 837,616,880 |
| Insertions | 8,865,838 |
| Mapped reads with at least one insertion | 16.46% |
| Deletions | 6,689,920 |
| Mapped reads with at least one deletion | 14.79% |
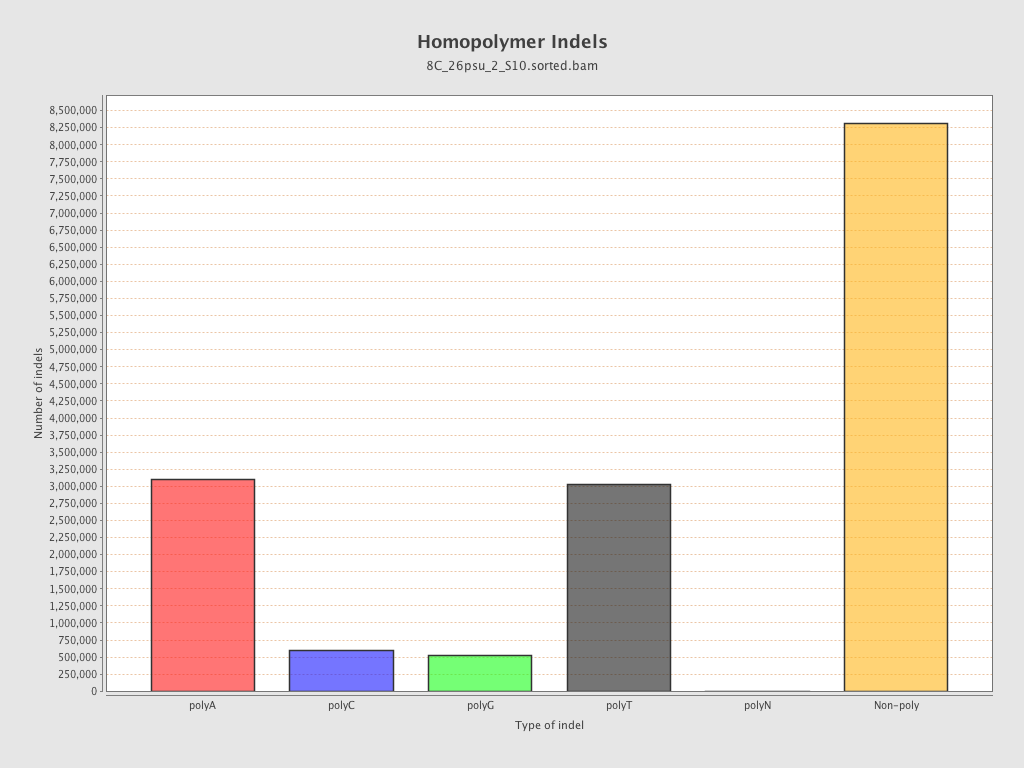
| Homopolymer indels | 46.59% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 305629318 | 1.9217 | 7.8957 |
| NC_027301.1 | 72943711 | 128627781 | 1.7634 | 6.2384 |
| NC_027302.1 | 92503428 | 165885993 | 1.7933 | 5.5324 |
| NC_027303.1 | 82398023 | 154330887 | 1.873 | 9.0182 |
| NC_027304.1 | 80503876 | 143076856 | 1.7773 | 7.5735 |
| NC_027305.1 | 87043187 | 153194338 | 1.76 | 11.1252 |
| NC_027306.1 | 58785265 | 106565244 | 1.8128 | 7.9671 |
| NC_027307.1 | 26434011 | 47324125 | 1.7903 | 11.4697 |
| NC_027308.1 | 141712163 | 259616060 | 1.832 | 6.9593 |
| NC_027309.1 | 116138521 | 225666999 | 1.9431 | 21.3667 |
| NC_027310.1 | 93888508 | 170123747 | 1.812 | 6.7698 |
| NC_027311.1 | 91880962 | 173339113 | 1.8866 | 11.9442 |
| NC_027312.1 | 107758822 | 205911073 | 1.9109 | 6.7895 |
| NC_027313.1 | 93901823 | 174278198 | 1.856 | 8.6273 |
| NC_027314.1 | 103963436 | 189213779 | 1.82 | 5.7316 |
| NC_027315.1 | 87796322 | 166432377 | 1.8957 | 8.6328 |
| NC_027316.1 | 57682537 | 102133353 | 1.7706 | 8.397 |
| NC_027317.1 | 70695409 | 135914854 | 1.9225 | 9.8424 |
| NC_027318.1 | 82978132 | 154235491 | 1.8587 | 27.6684 |
| NC_027319.1 | 86795997 | 161458667 | 1.8602 | 7.5533 |
| NC_027320.1 | 58021487 | 102066498 | 1.7591 | 5.1711 |
| NC_027321.1 | 63420196 | 124844674 | 1.9685 | 22.5712 |
| NC_027322.1 | 49854004 | 93011580 | 1.8657 | 7.21 |
| NC_027323.1 | 48650976 | 84956928 | 1.7463 | 6.2368 |
| NC_027324.1 | 51481326 | 93835373 | 1.8227 | 6.1213 |
| NC_027325.1 | 47900953 | 83114816 | 1.7351 | 8.7425 |
| NC_027326.1 | 43943985 | 79044929 | 1.7988 | 7.3158 |
| NC_027327.1 | 39600944 | 73697890 | 1.861 | 13.1615 |
| NC_027328.1 | 42488238 | 71561653 | 1.6843 | 9.3021 |
| NC_001960.1 | 16665 | 18402032 | 1,104.2323 | 1,676.515 |