Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 31,221,158 |
| Mapped reads | 31,221,158 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 31,221,158 / 100% |
| Mapped reads, first in pair | 15,610,579 / 50% |
| Mapped reads, second in pair | 15,610,579 / 50% |
| Mapped reads, both in pair | 31,221,158 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 117.1 |
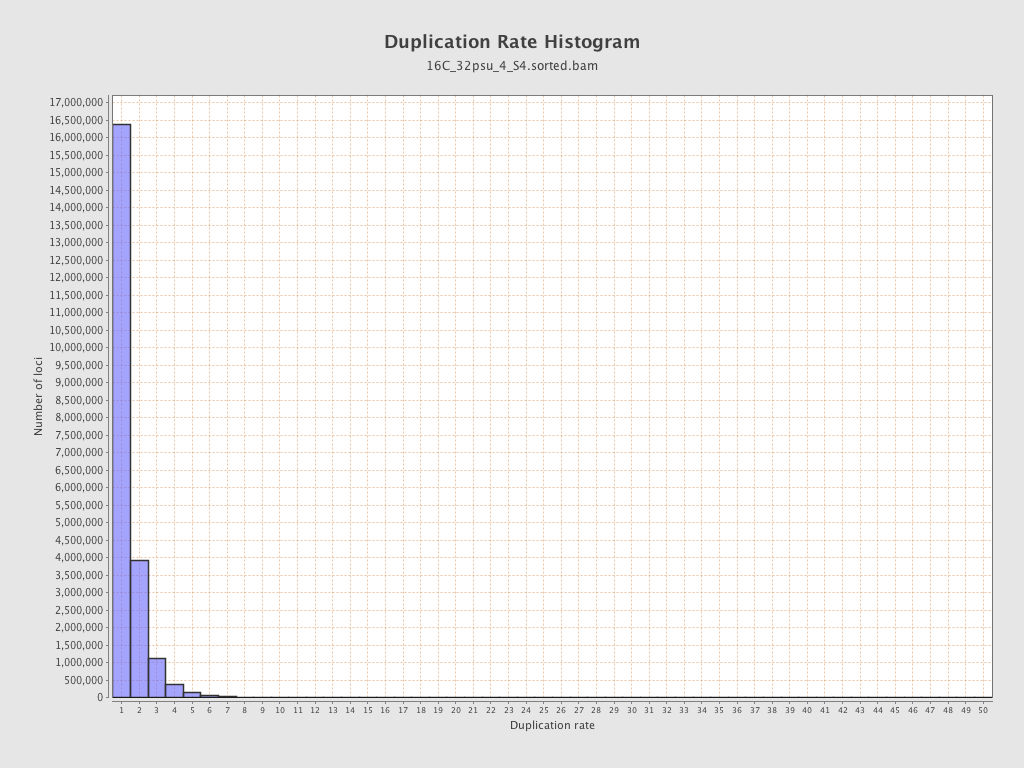
| Duplicated reads (estimated) | 9,153,913 / 29.32% |
| Duplication rate | 25.74% |
| Clipped reads | 0 / 0% |
ACGT Content
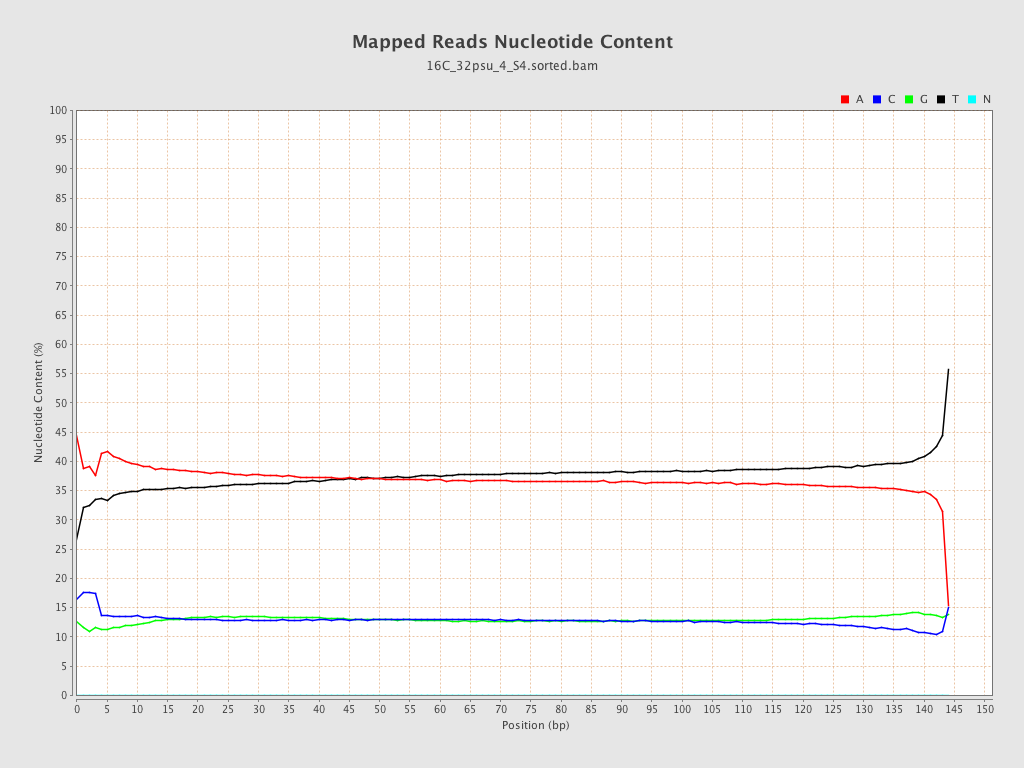
| Number/percentage of A's | 1,347,847,288 / 37.13% |
| Number/percentage of C's | 468,069,404 / 12.9% |
| Number/percentage of T's | 1,347,193,224 / 37.12% |
| Number/percentage of G's | 466,653,441 / 12.86% |
| Number/percentage of N's | 25,556 / 0% |
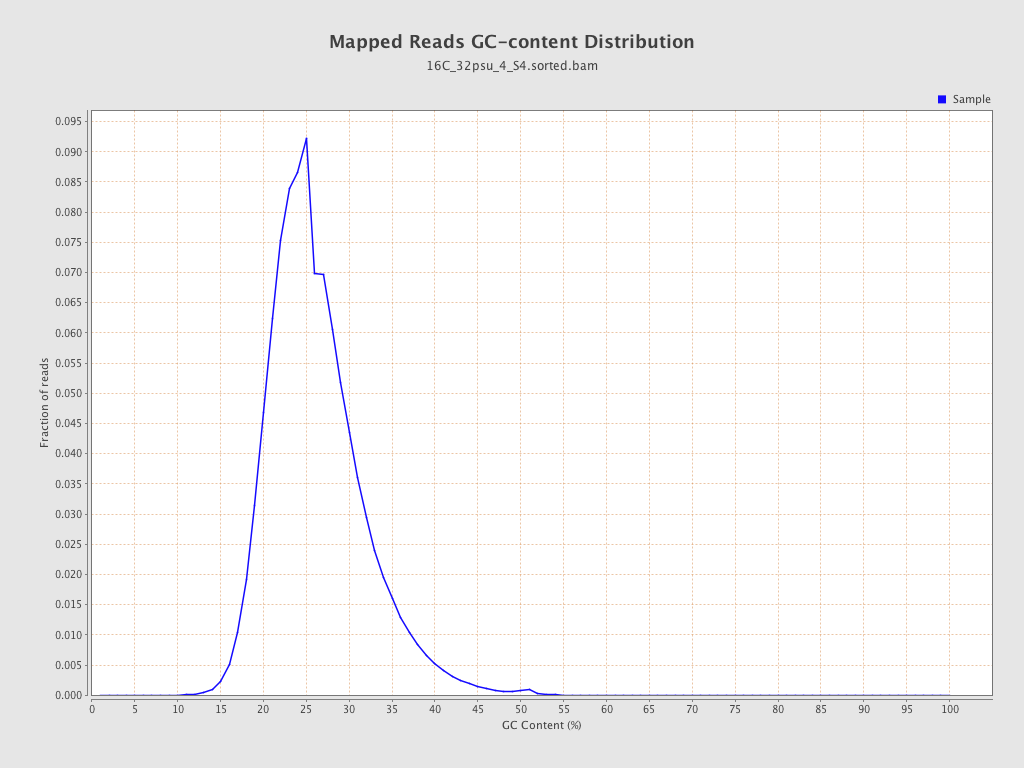
| GC Percentage | 25.75% |
Coverage
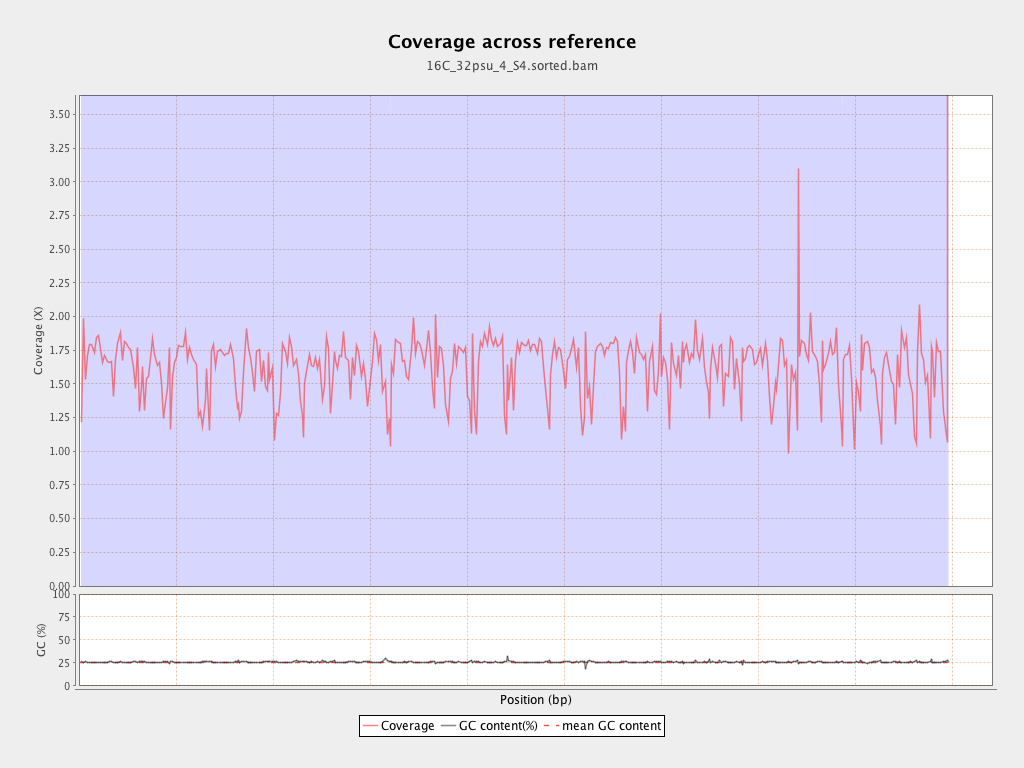
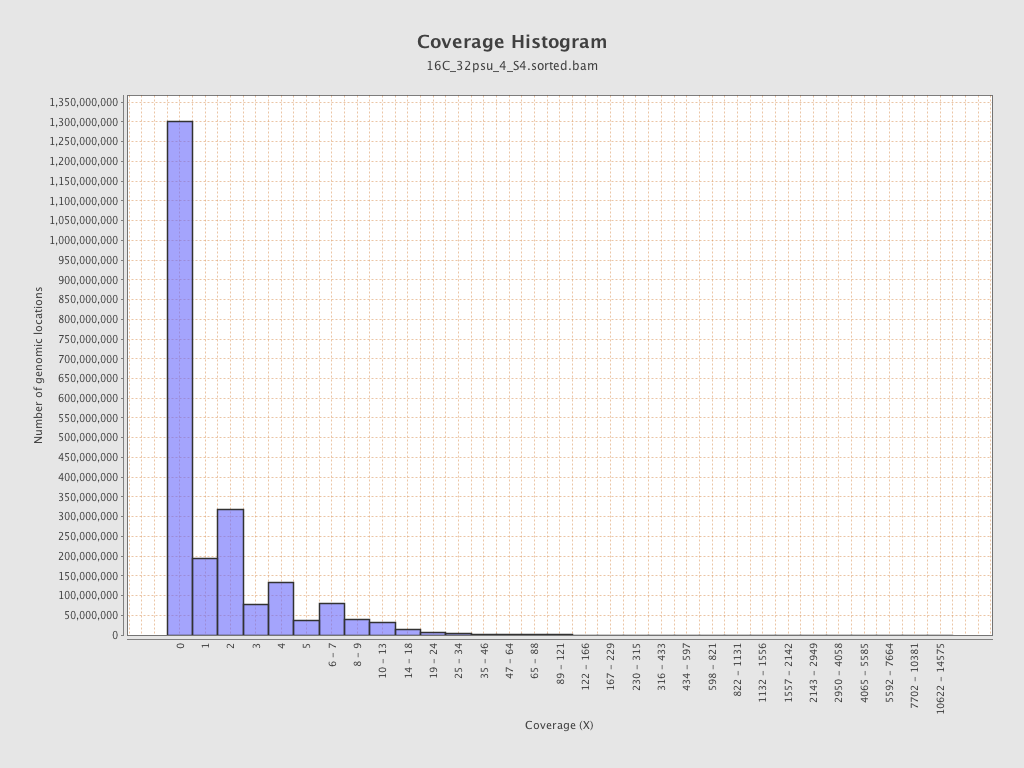
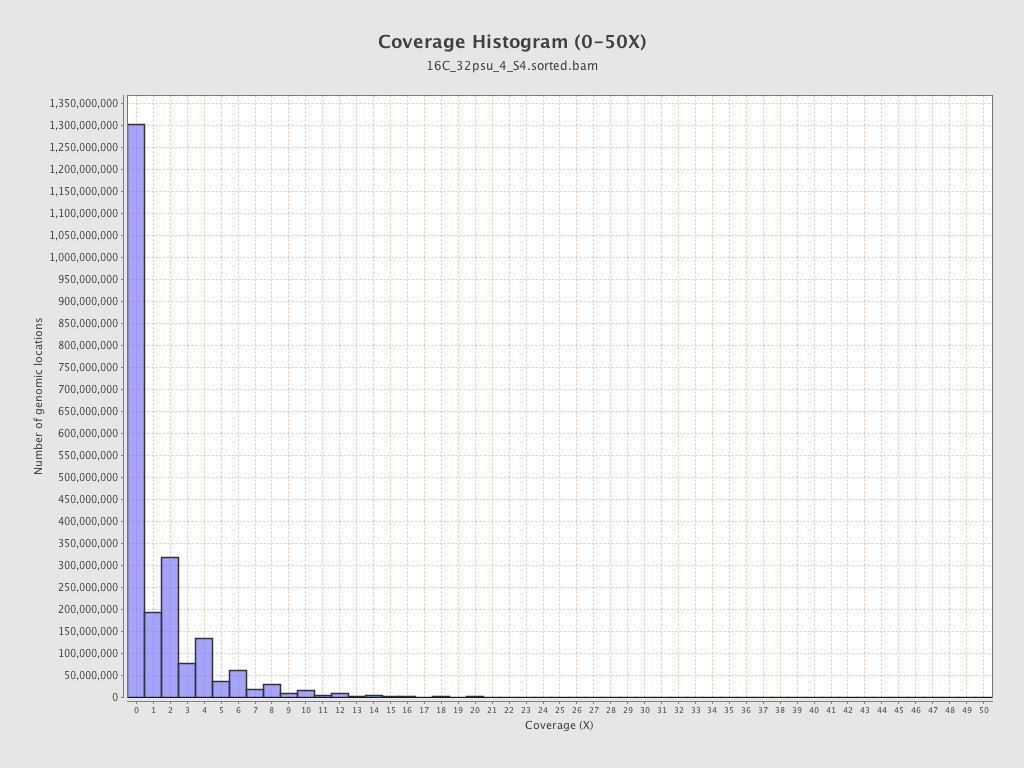
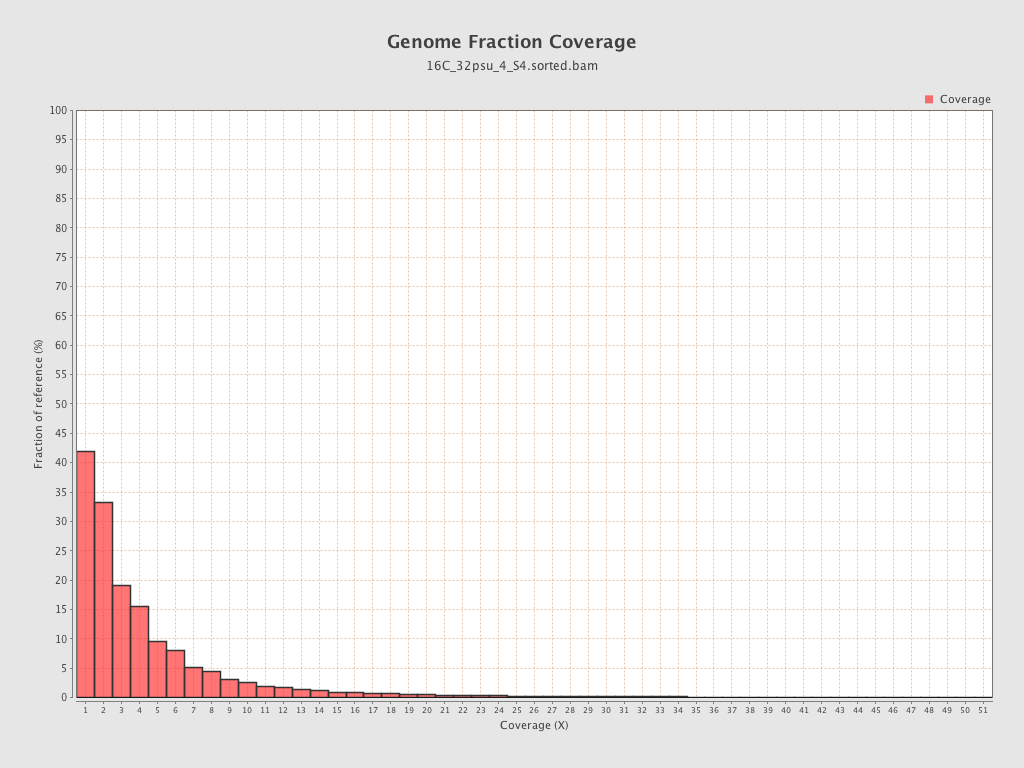
| Mean | 1.6248 |
| Standard Deviation | 8.5179 |
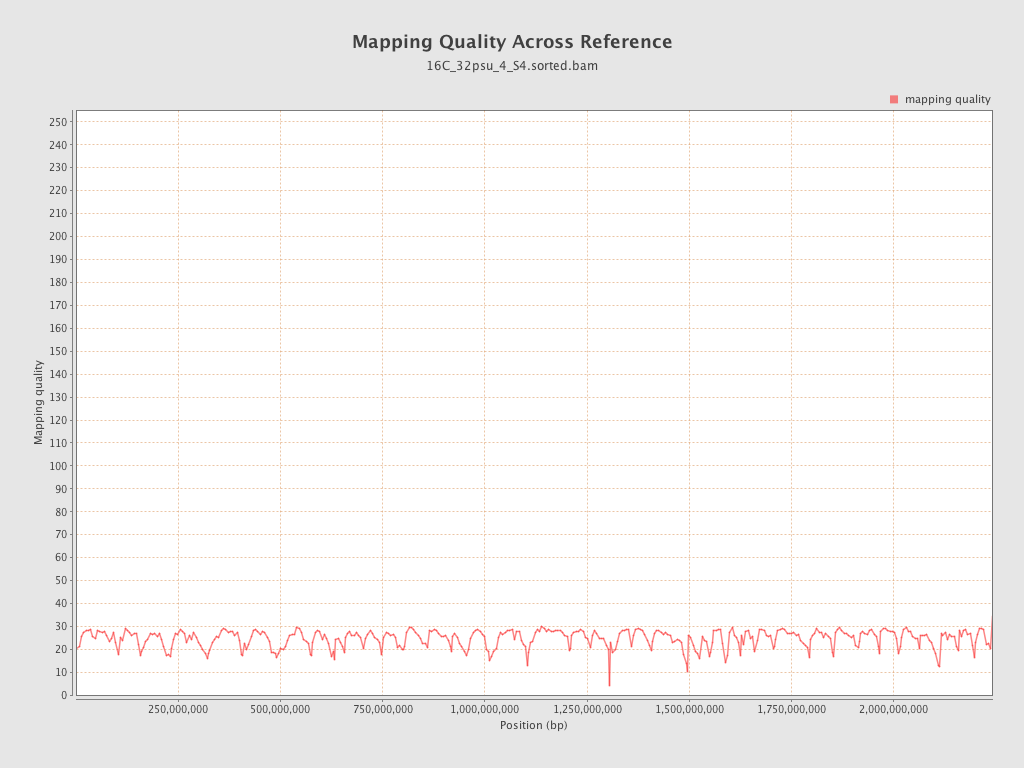
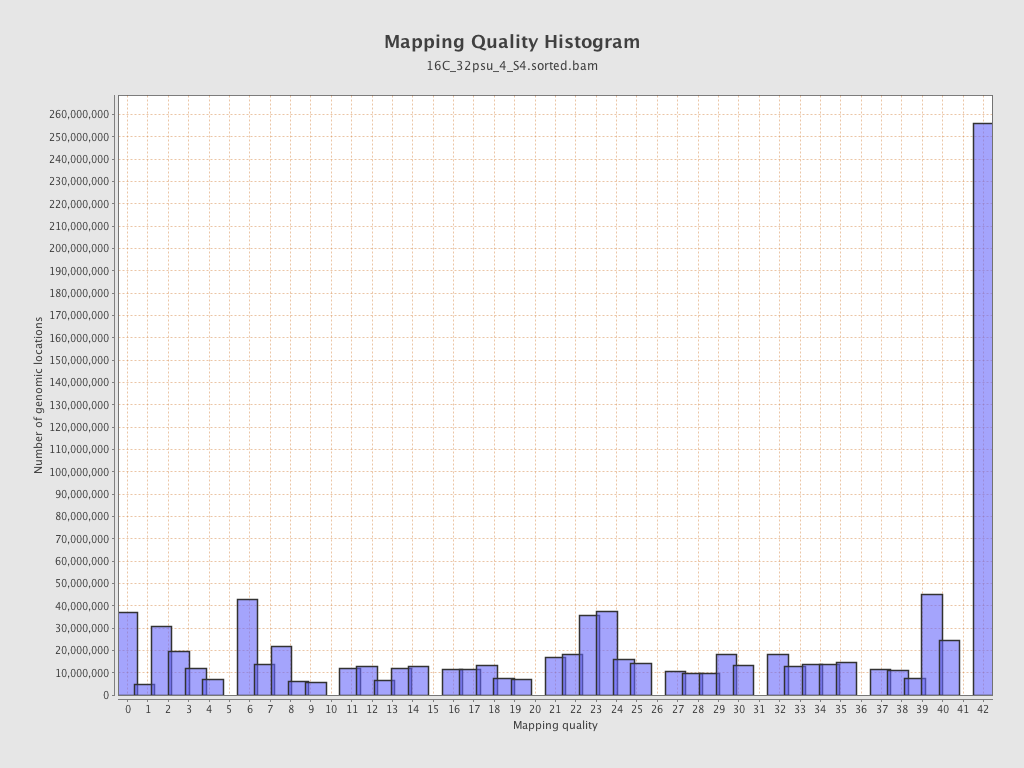
Mapping Quality
| Mean Mapping Quality | 24.51 |
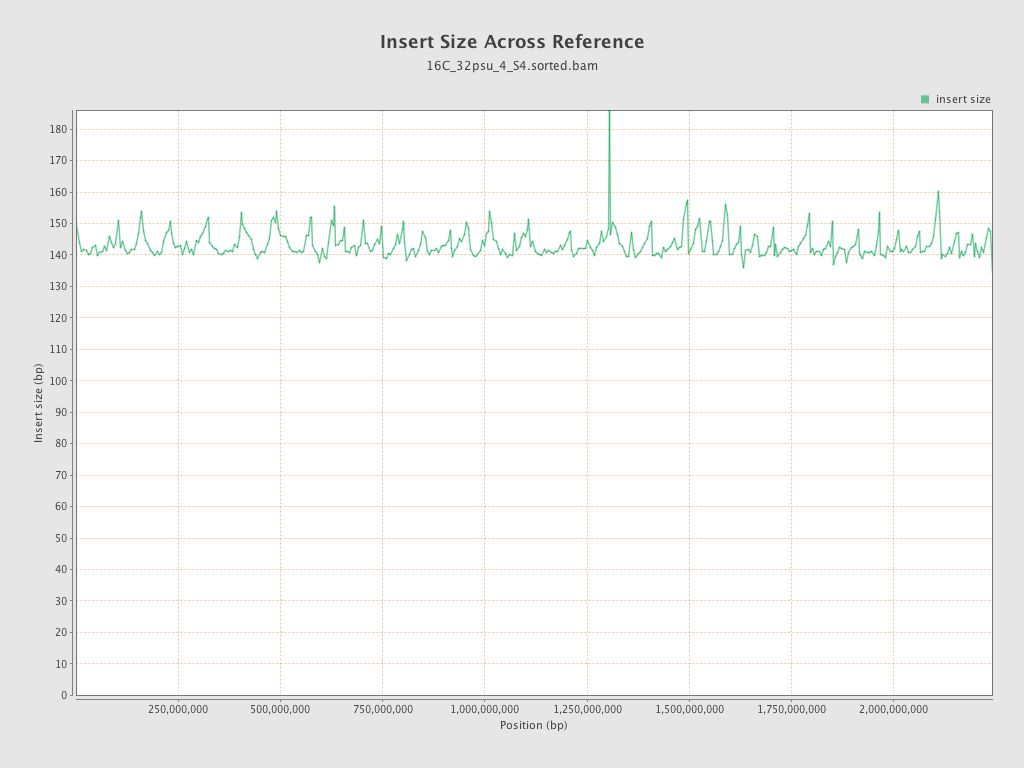
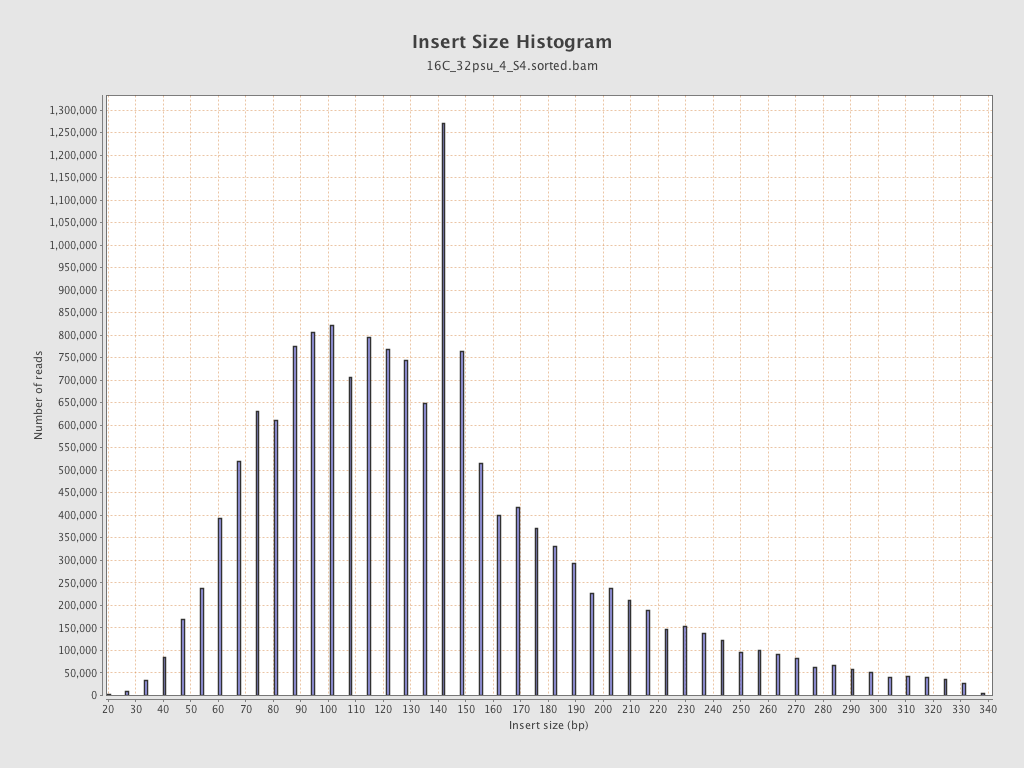
Insert size
| Mean | 143.51 |
| Standard Deviation | 65.95 |
| P25/Median/P75 | 98 / 133 / 169 |
Mismatches and indels
| General error rate | 21.09% |
| Mismatches | 740,419,680 |
| Insertions | 8,441,149 |
| Mapped reads with at least one insertion | 18.94% |
| Deletions | 6,089,037 |
| Mapped reads with at least one deletion | 16.62% |
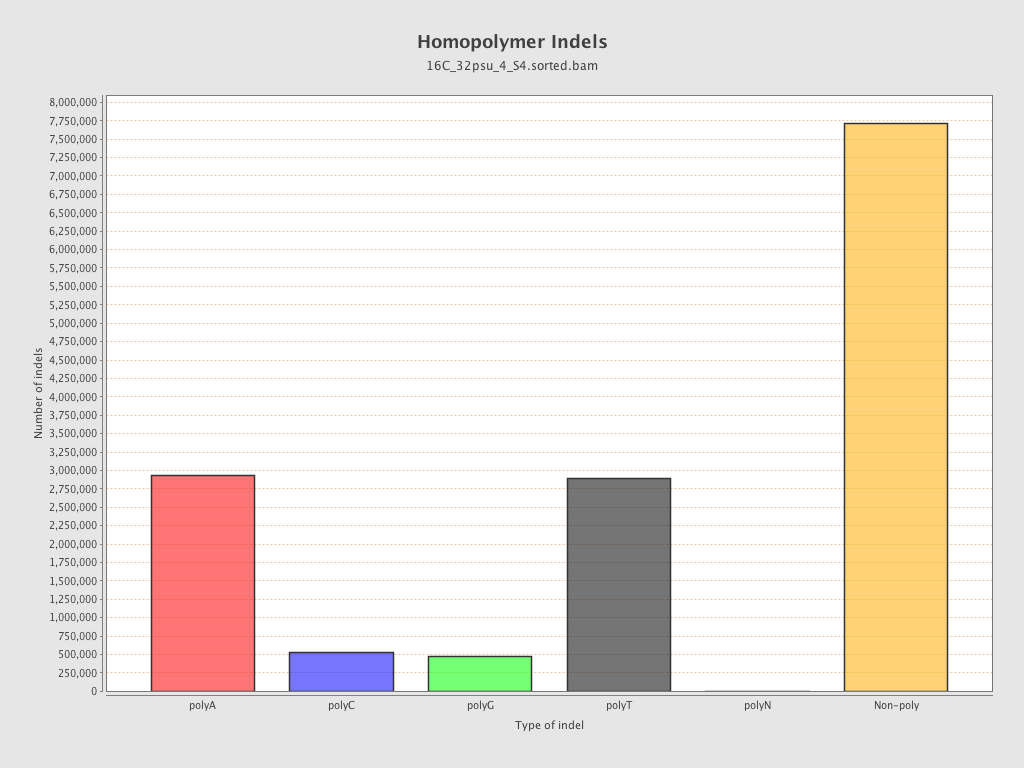
| Homopolymer indels | 46.94% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 268401169 | 1.6876 | 6.2497 |
| NC_027301.1 | 72943711 | 113315447 | 1.5535 | 4.9002 |
| NC_027302.1 | 92503428 | 147003245 | 1.5892 | 4.8338 |
| NC_027303.1 | 82398023 | 135834227 | 1.6485 | 5.7926 |
| NC_027304.1 | 80503876 | 126961687 | 1.5771 | 6.3906 |
| NC_027305.1 | 87043187 | 134220264 | 1.542 | 8.6961 |
| NC_027306.1 | 58785265 | 92508387 | 1.5737 | 5.9037 |
| NC_027307.1 | 26434011 | 42812568 | 1.6196 | 9.5253 |
| NC_027308.1 | 141712163 | 227427376 | 1.6049 | 5.7219 |
| NC_027309.1 | 116138521 | 198035188 | 1.7052 | 16.4875 |
| NC_027310.1 | 93888508 | 149528077 | 1.5926 | 5.3407 |
| NC_027311.1 | 91880962 | 152945097 | 1.6646 | 8.5531 |
| NC_027312.1 | 107758822 | 180379884 | 1.6739 | 5.3188 |
| NC_027313.1 | 93901823 | 151790426 | 1.6165 | 6.3125 |
| NC_027314.1 | 103963436 | 166926155 | 1.6056 | 4.5264 |
| NC_027315.1 | 87796322 | 145489074 | 1.6571 | 6.547 |
| NC_027316.1 | 57682537 | 91913017 | 1.5934 | 6.9048 |
| NC_027317.1 | 70695409 | 119848236 | 1.6953 | 7.6346 |
| NC_027318.1 | 82978132 | 133510071 | 1.609 | 13.7947 |
| NC_027319.1 | 86795997 | 141374048 | 1.6288 | 5.4079 |
| NC_027320.1 | 58021487 | 90031003 | 1.5517 | 4.1151 |
| NC_027321.1 | 63420196 | 108376594 | 1.7089 | 13.0418 |
| NC_027322.1 | 49854004 | 81980667 | 1.6444 | 5.5683 |
| NC_027323.1 | 48650976 | 74369864 | 1.5286 | 4.7906 |
| NC_027324.1 | 51481326 | 82173418 | 1.5962 | 5.0435 |
| NC_027325.1 | 47900953 | 74829565 | 1.5622 | 7.1517 |
| NC_027326.1 | 43943985 | 69018837 | 1.5706 | 5.1021 |
| NC_027327.1 | 39600944 | 64656495 | 1.6327 | 9.7002 |
| NC_027328.1 | 42488238 | 62988955 | 1.4825 | 8.0735 |
| NC_001960.1 | 16665 | 11156163 | 669.4367 | 1,056.6341 |