Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 38,429,850 |
| Mapped reads | 38,429,850 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 38,429,850 / 100% |
| Mapped reads, first in pair | 19,214,925 / 50% |
| Mapped reads, second in pair | 19,214,925 / 50% |
| Mapped reads, both in pair | 38,429,850 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 103.51 |
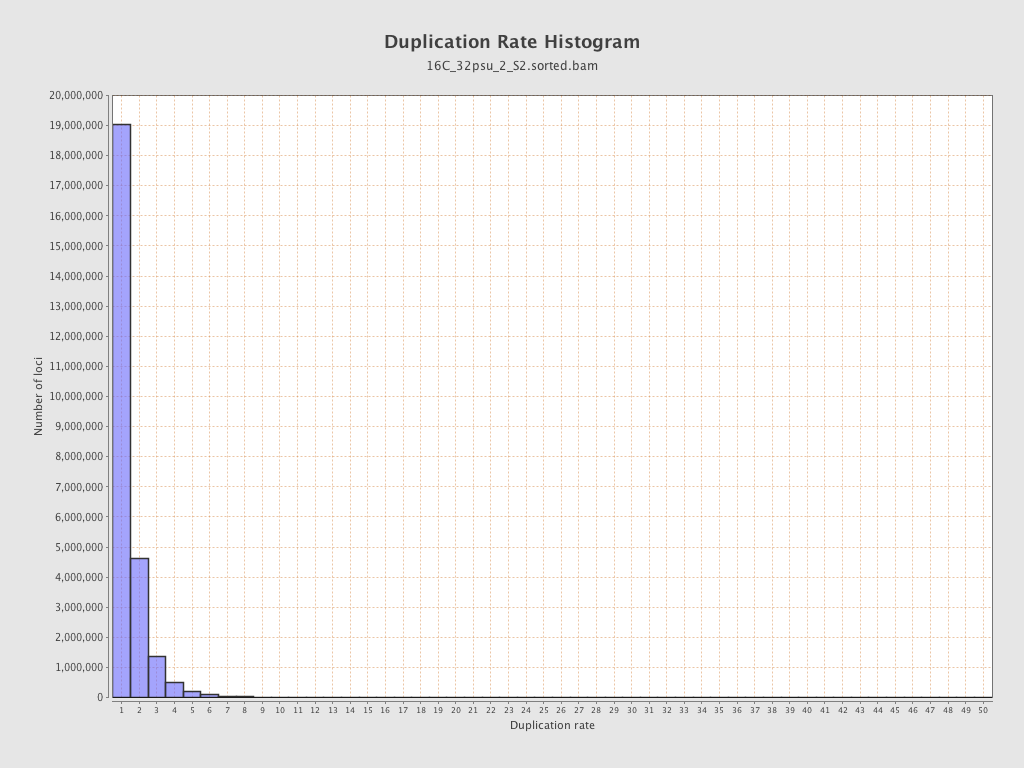
| Duplicated reads (estimated) | 12,418,542 / 32.31% |
| Duplication rate | 26.77% |
| Clipped reads | 0 / 0% |
ACGT Content
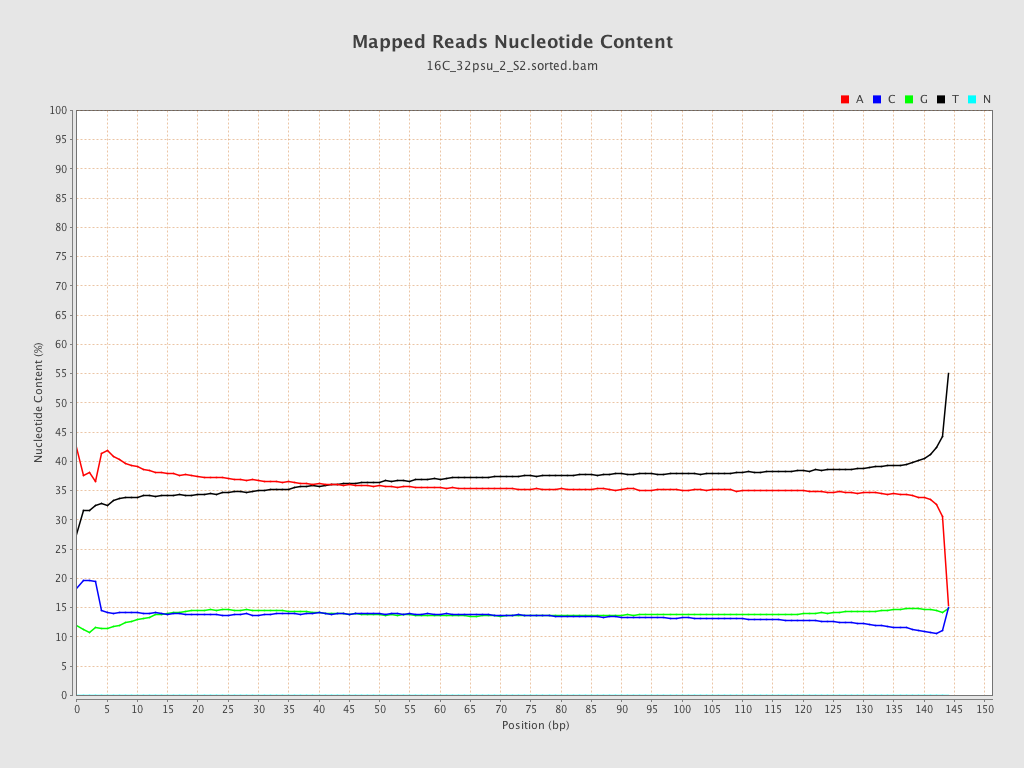
| Number/percentage of A's | 1,432,176,979 / 36.25% |
| Number/percentage of C's | 546,236,321 / 13.83% |
| Number/percentage of T's | 1,429,811,033 / 36.19% |
| Number/percentage of G's | 542,478,830 / 13.73% |
| Number/percentage of N's | 24,652 / 0% |
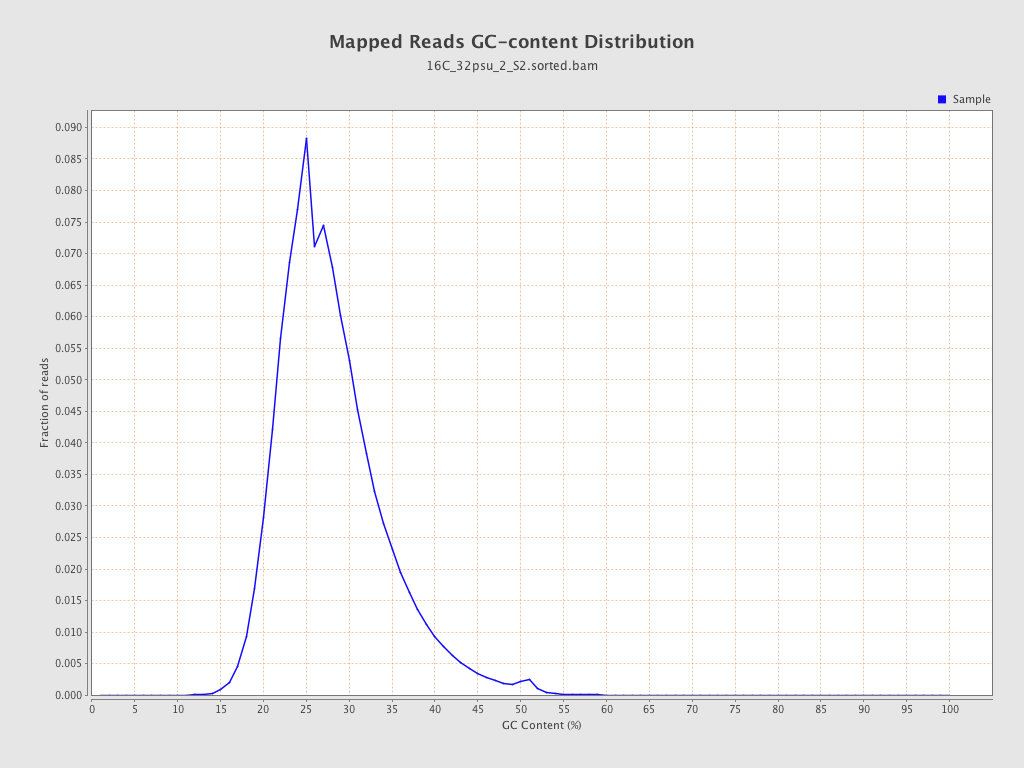
| GC Percentage | 27.56% |
Coverage
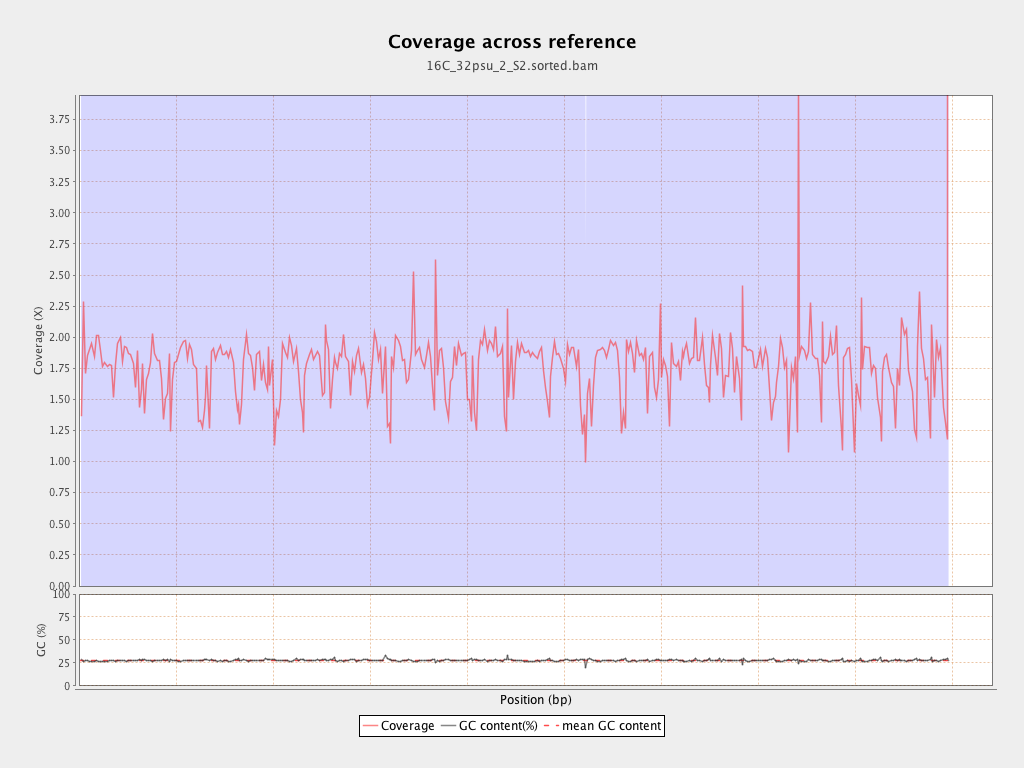
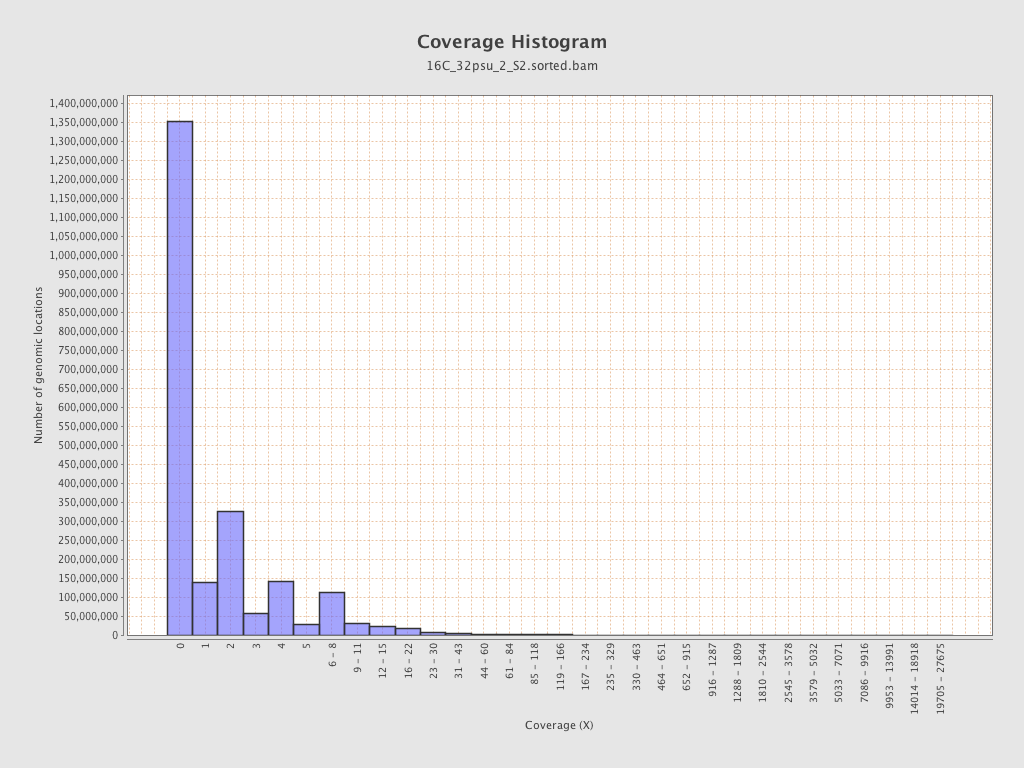
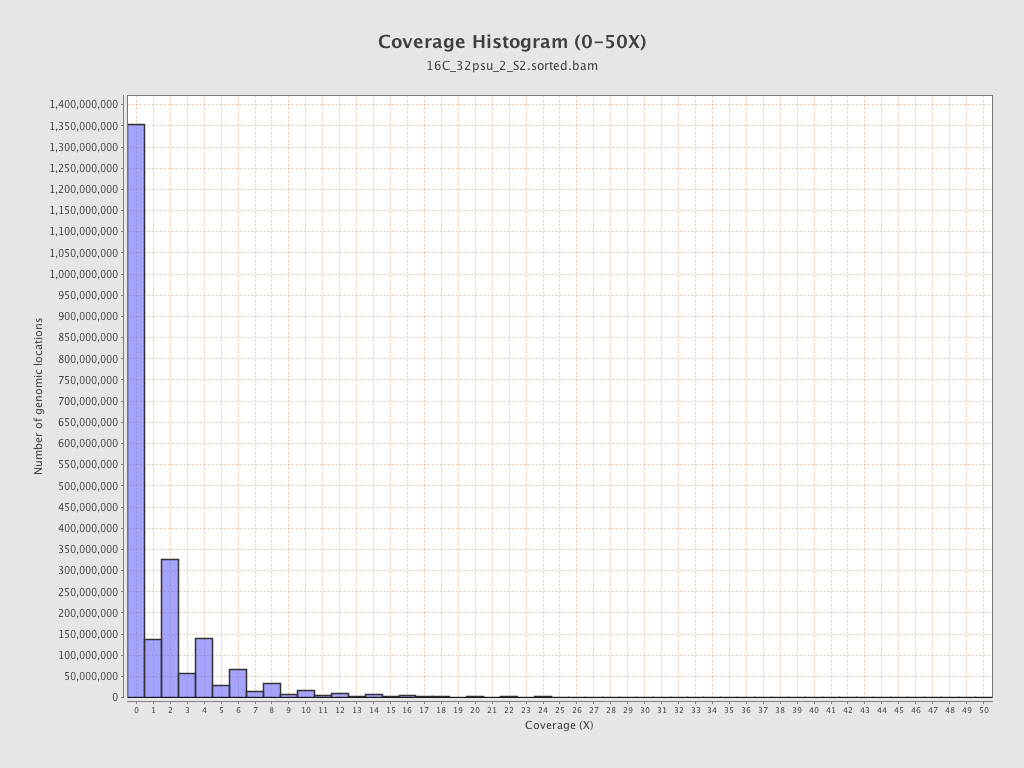
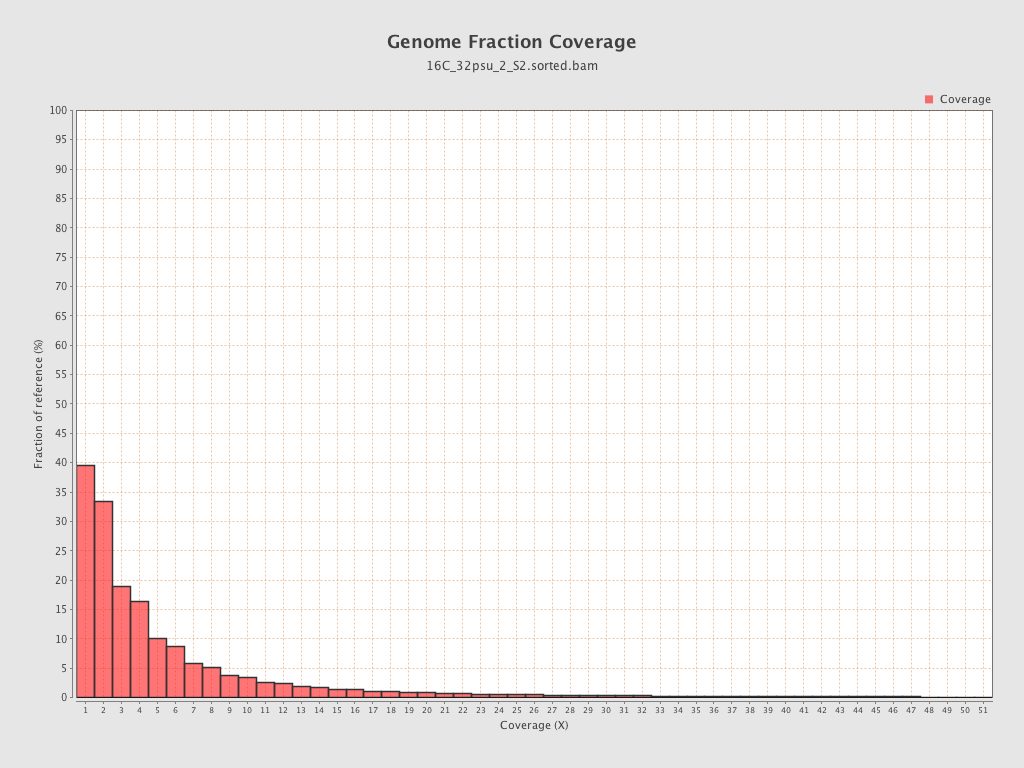
| Mean | 1.7683 |
| Standard Deviation | 13.8162 |
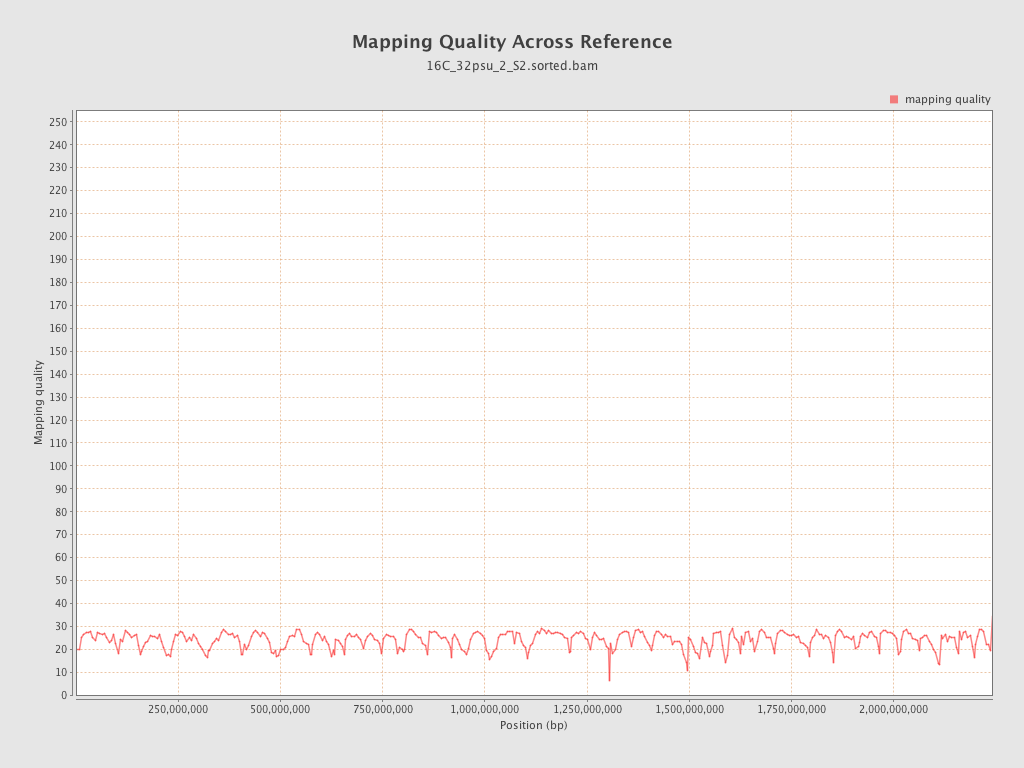
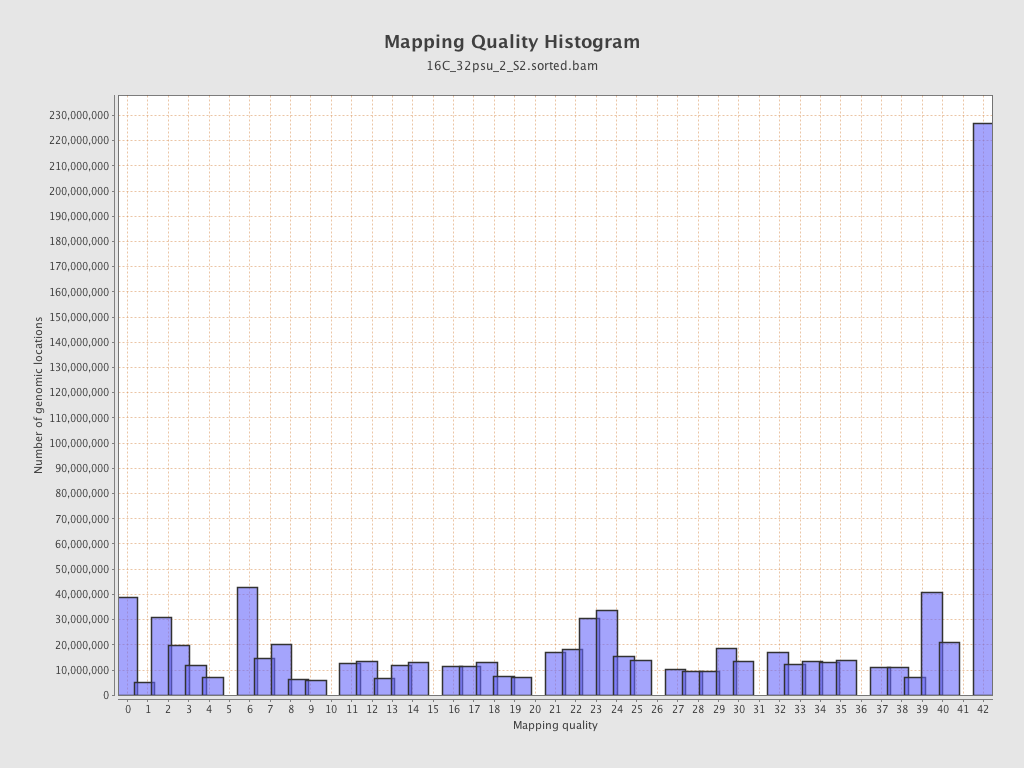
Mapping Quality
| Mean Mapping Quality | 23.91 |
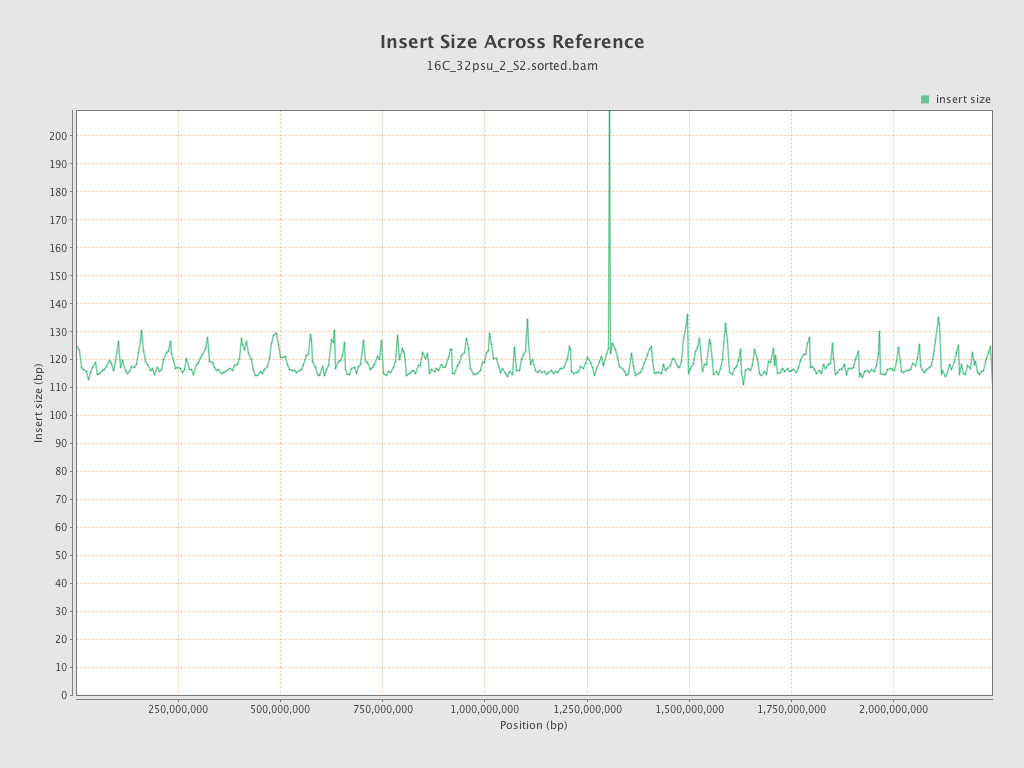
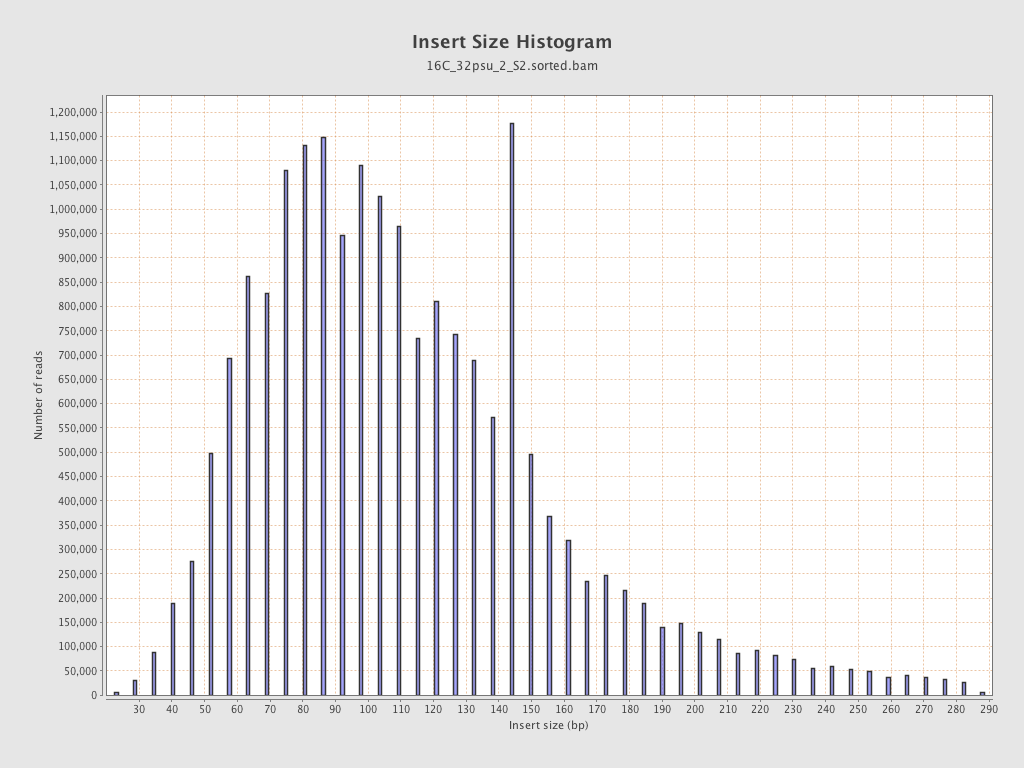
Insert size
| Mean | 118.68 |
| Standard Deviation | 54.99 |
| P25/Median/P75 | 82 / 108 / 144 |
Mismatches and indels
| General error rate | 20.64% |
| Mismatches | 789,112,360 |
| Insertions | 9,083,145 |
| Mapped reads with at least one insertion | 17.6% |
| Deletions | 6,471,563 |
| Mapped reads with at least one deletion | 14.52% |
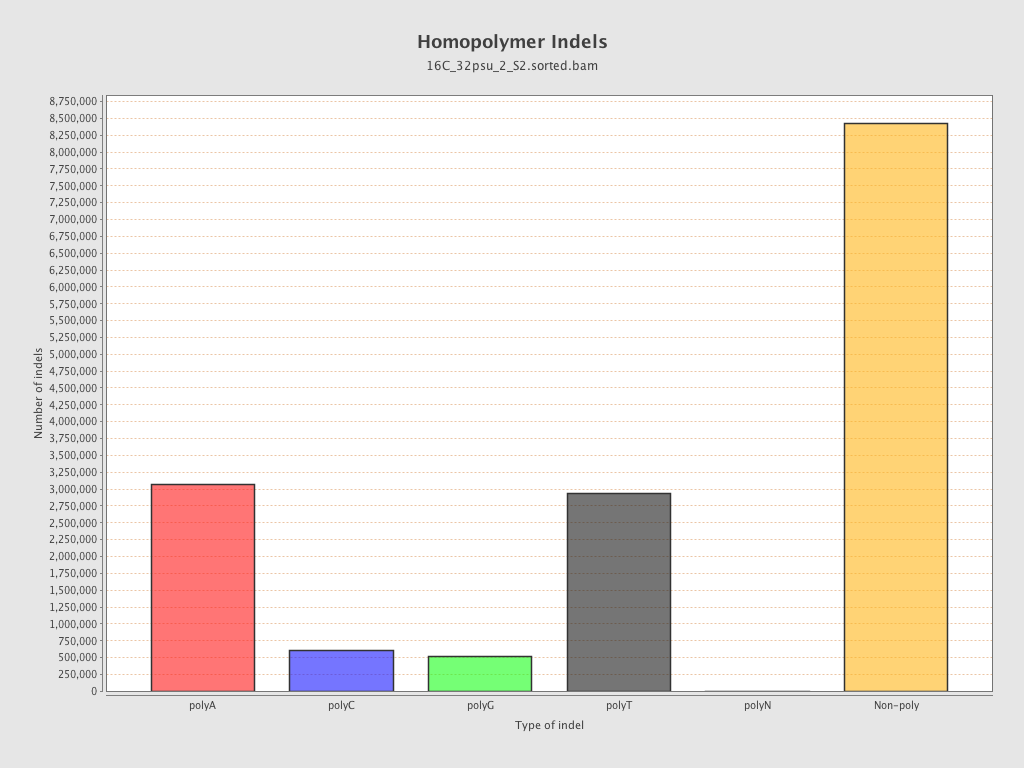
| Homopolymer indels | 45.86% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 289425468 | 1.8198 | 8.0366 |
| NC_027301.1 | 72943711 | 123249000 | 1.6896 | 6.4594 |
| NC_027302.1 | 92503428 | 157713897 | 1.705 | 5.4777 |
| NC_027303.1 | 82398023 | 146619640 | 1.7794 | 8.4386 |
| NC_027304.1 | 80503876 | 137834549 | 1.7121 | 9.0302 |
| NC_027305.1 | 87043187 | 146157345 | 1.6791 | 10.6696 |
| NC_027306.1 | 58785265 | 103115710 | 1.7541 | 8.937 |
| NC_027307.1 | 26434011 | 46382808 | 1.7547 | 15.7243 |
| NC_027308.1 | 141712163 | 247513233 | 1.7466 | 8.8548 |
| NC_027309.1 | 116138521 | 214893762 | 1.8503 | 30.2447 |
| NC_027310.1 | 93888508 | 161810845 | 1.7234 | 6.6835 |
| NC_027311.1 | 91880962 | 163878923 | 1.7836 | 12.6057 |
| NC_027312.1 | 107758822 | 193250333 | 1.7934 | 7.6764 |
| NC_027313.1 | 93901823 | 164751582 | 1.7545 | 8.4477 |
| NC_027314.1 | 103963436 | 179534321 | 1.7269 | 5.6867 |
| NC_027315.1 | 87796322 | 157722485 | 1.7965 | 8.9325 |
| NC_027316.1 | 57682537 | 99806142 | 1.7303 | 8.9238 |
| NC_027317.1 | 70695409 | 129736415 | 1.8351 | 9.8404 |
| NC_027318.1 | 82978132 | 146699330 | 1.7679 | 23.4108 |
| NC_027319.1 | 86795997 | 153787134 | 1.7718 | 8.5923 |
| NC_027320.1 | 58021487 | 97820901 | 1.6859 | 5.3606 |
| NC_027321.1 | 63420196 | 118694248 | 1.8716 | 19.7204 |
| NC_027322.1 | 49854004 | 89170940 | 1.7886 | 7.7284 |
| NC_027323.1 | 48650976 | 80606521 | 1.6568 | 6.1768 |
| NC_027324.1 | 51481326 | 90208913 | 1.7523 | 8.3094 |
| NC_027325.1 | 47900953 | 80126334 | 1.6728 | 7.2734 |
| NC_027326.1 | 43943985 | 77183241 | 1.7564 | 8.3294 |
| NC_027327.1 | 39600944 | 71465046 | 1.8046 | 12.4363 |
| NC_027328.1 | 42488238 | 68955185 | 1.6229 | 9.967 |
| NC_001960.1 | 16665 | 23307248 | 1,398.5747 | 2,079.6292 |