Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 29,590,976 |
| Mapped reads | 29,590,976 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 29,590,976 / 100% |
| Mapped reads, first in pair | 14,795,488 / 50% |
| Mapped reads, second in pair | 14,795,488 / 50% |
| Mapped reads, both in pair | 29,590,976 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 112.99 |
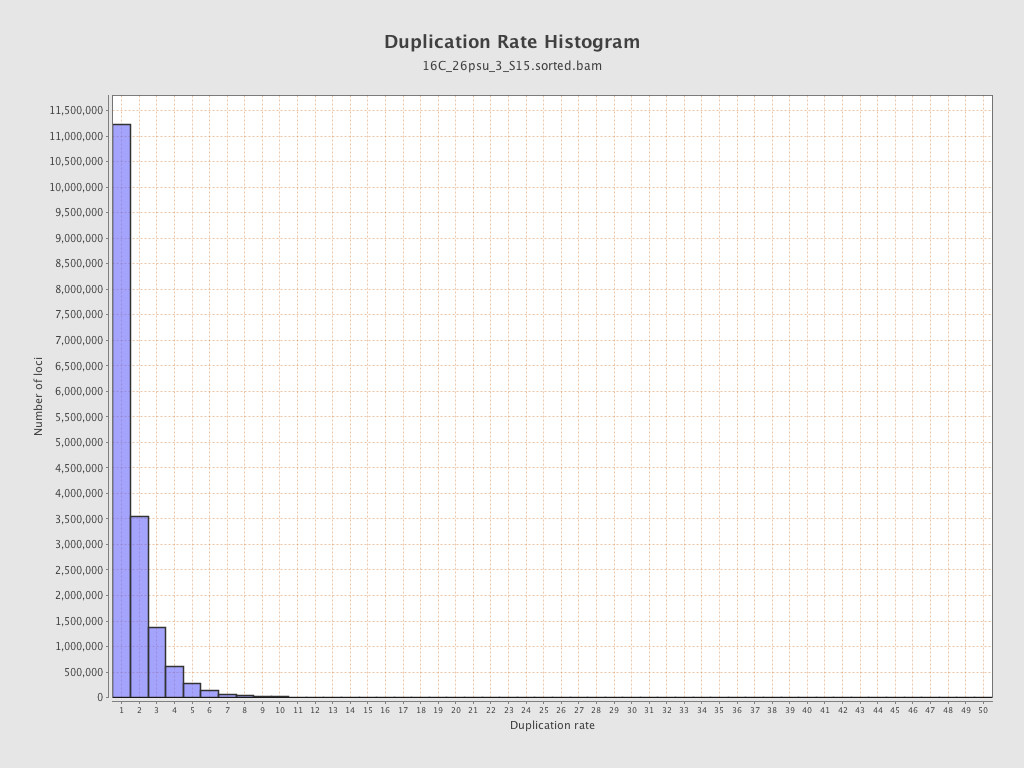
| Duplicated reads (estimated) | 12,217,026 / 41.29% |
| Duplication rate | 35.33% |
| Clipped reads | 0 / 0% |
ACGT Content
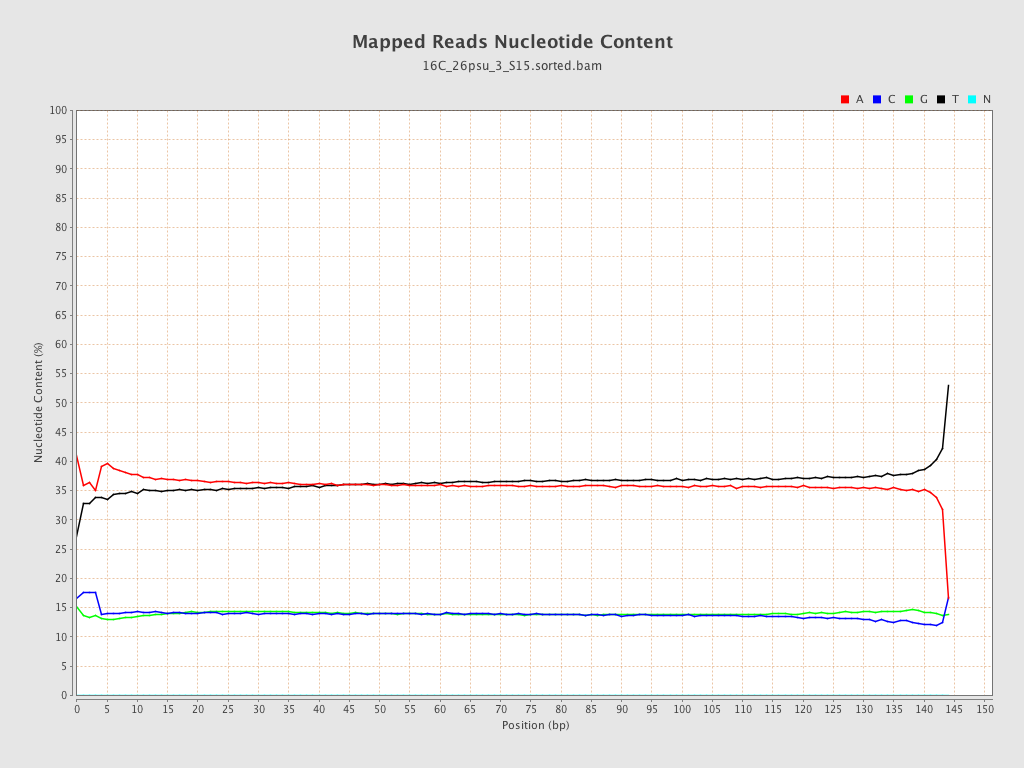
| Number/percentage of A's | 1,200,312,514 / 36.12% |
| Number/percentage of C's | 462,029,408 / 13.9% |
| Number/percentage of T's | 1,197,858,197 / 36.05% |
| Number/percentage of G's | 462,709,664 / 13.92% |
| Number/percentage of N's | 22,557 / 0% |
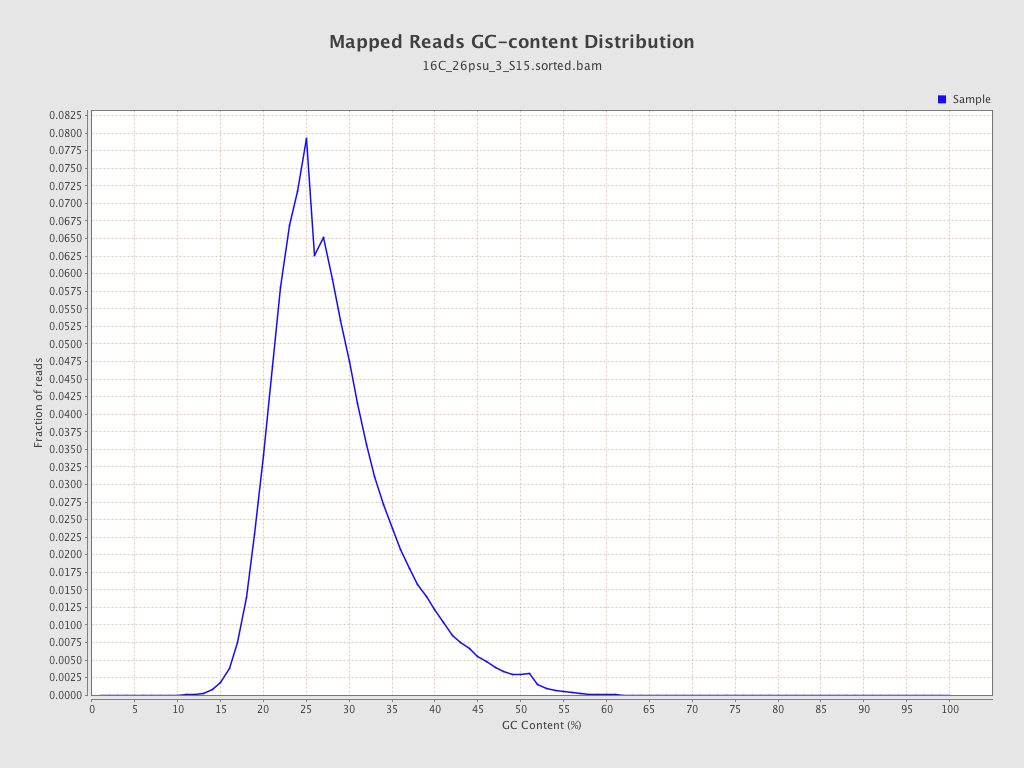
| GC Percentage | 27.83% |
Coverage
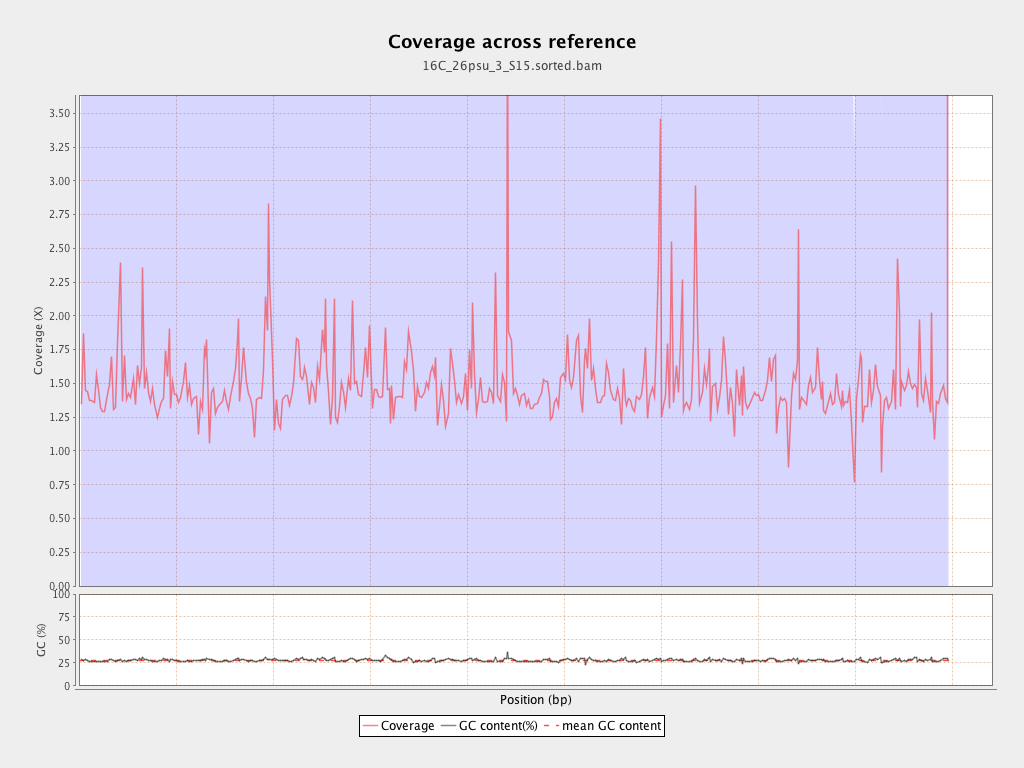
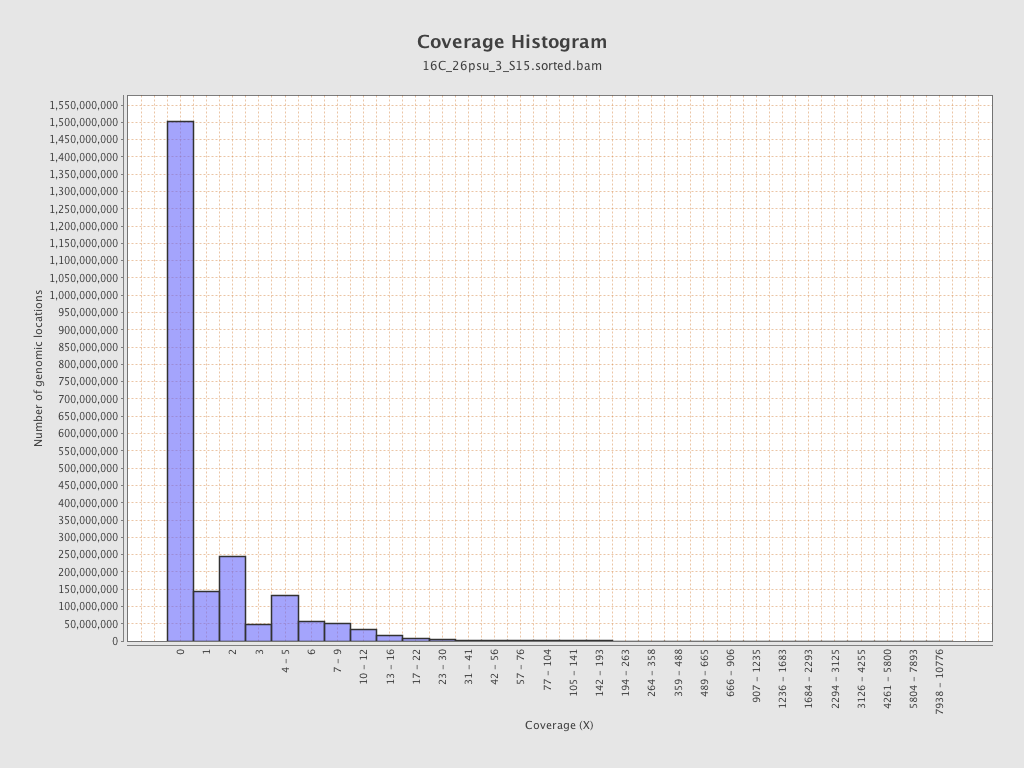
| Mean | 1.4876 |
| Standard Deviation | 11.1563 |
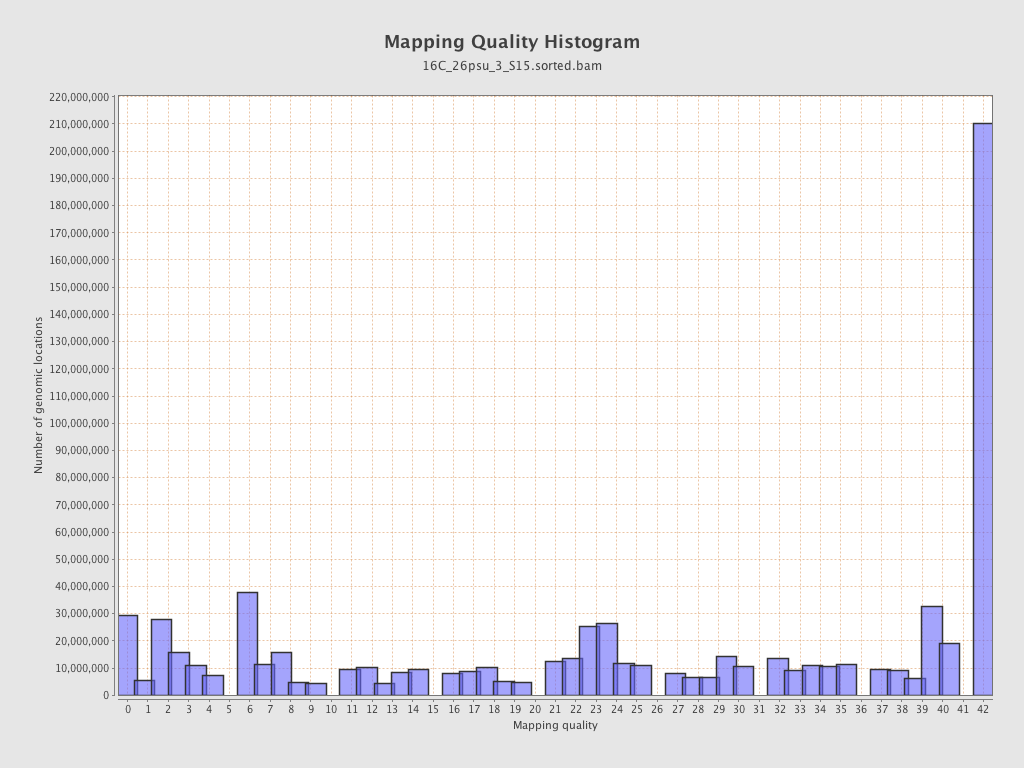
Mapping Quality
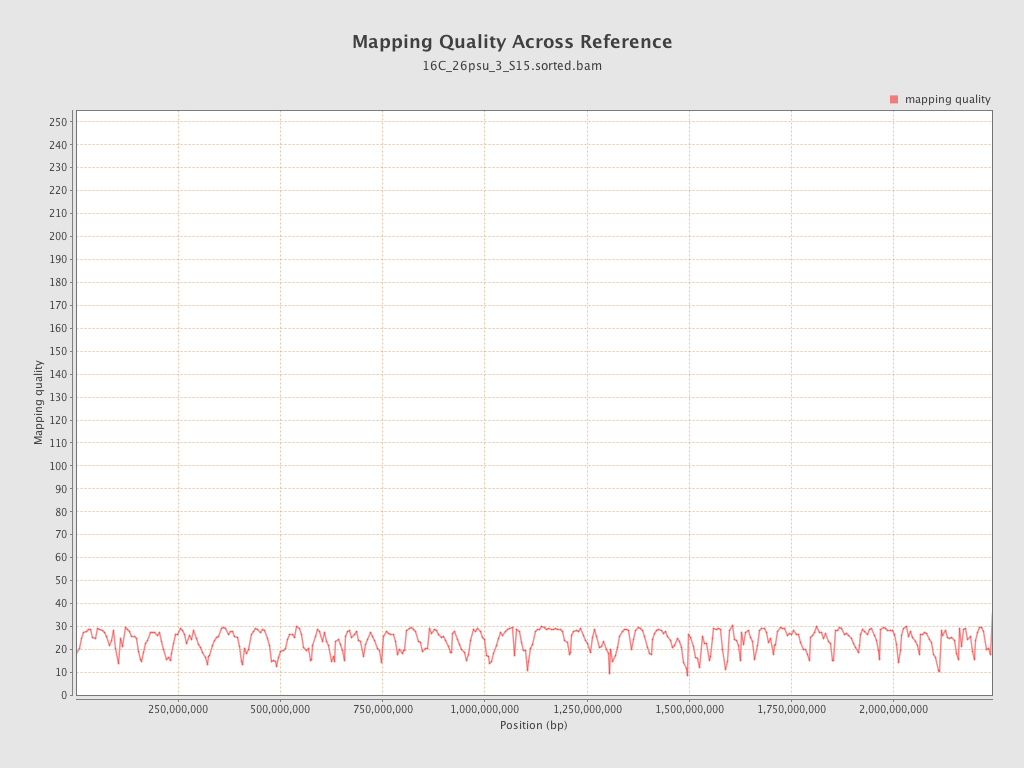
| Mean Mapping Quality | 23.67 |
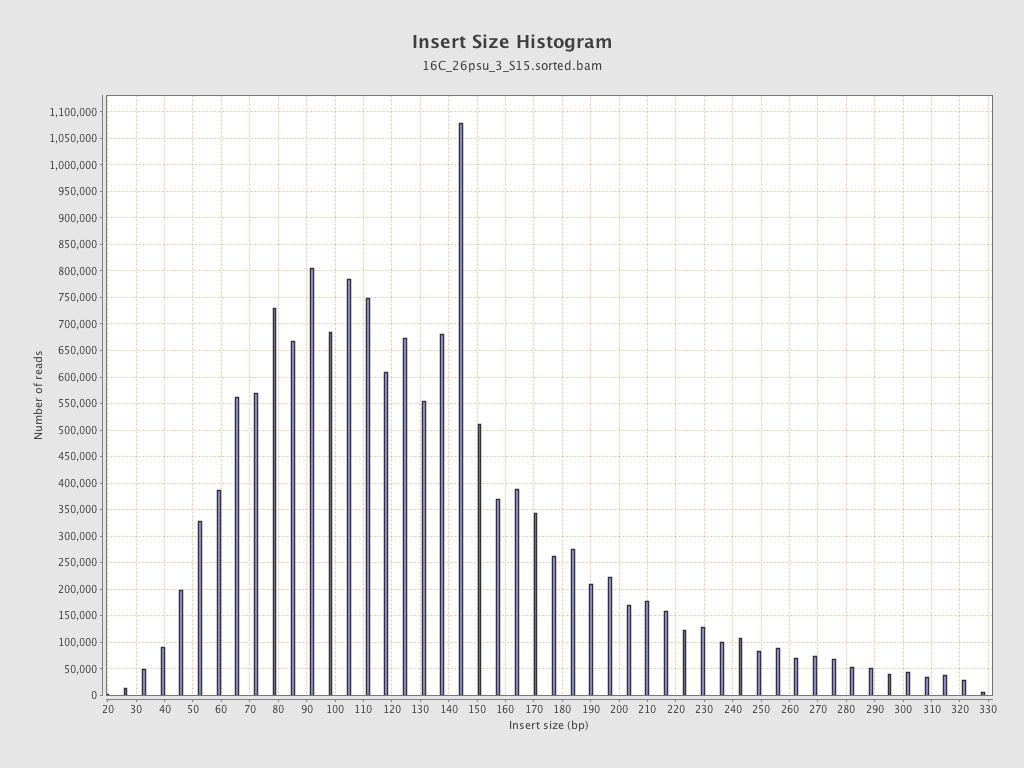
Insert size
| Mean | 139.68 |
| Standard Deviation | 69.65 |
| P25/Median/P75 | 92 / 126 / 164 |
Mismatches and indels
| General error rate | 19.58% |
| Mismatches | 630,724,283 |
| Insertions | 6,942,947 |
| Mapped reads with at least one insertion | 16.63% |
| Deletions | 5,591,620 |
| Mapped reads with at least one deletion | 15.9% |
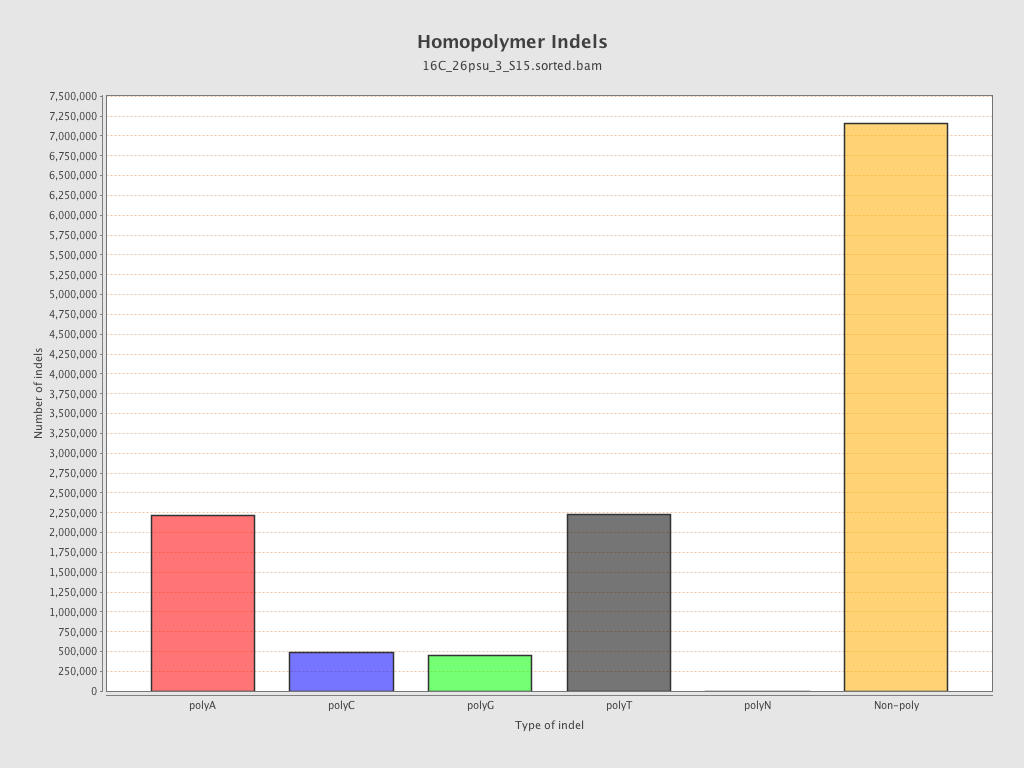
| Homopolymer indels | 42.93% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 238119142 | 1.4972 | 10.428 |
| NC_027301.1 | 72943711 | 109117144 | 1.4959 | 9.752 |
| NC_027302.1 | 92503428 | 131534443 | 1.4219 | 7.2226 |
| NC_027303.1 | 82398023 | 117869589 | 1.4305 | 6.8961 |
| NC_027304.1 | 80503876 | 123150455 | 1.5297 | 8.871 |
| NC_027305.1 | 87043187 | 130595123 | 1.5003 | 12.4419 |
| NC_027306.1 | 58785265 | 92065259 | 1.5661 | 8.8992 |
| NC_027307.1 | 26434011 | 40497664 | 1.532 | 14.0584 |
| NC_027308.1 | 141712163 | 213922902 | 1.5096 | 9.3282 |
| NC_027309.1 | 116138521 | 174312210 | 1.5009 | 15.037 |
| NC_027310.1 | 93888508 | 134906076 | 1.4369 | 9.4432 |
| NC_027311.1 | 91880962 | 136484399 | 1.4854 | 15.2228 |
| NC_027312.1 | 107758822 | 156643088 | 1.4536 | 7.8313 |
| NC_027313.1 | 93901823 | 142188133 | 1.5142 | 13.3342 |
| NC_027314.1 | 103963436 | 154552377 | 1.4866 | 9.793 |
| NC_027315.1 | 87796322 | 134609998 | 1.5332 | 12.1918 |
| NC_027316.1 | 57682537 | 94047355 | 1.6304 | 13.2249 |
| NC_027317.1 | 70695409 | 115015407 | 1.6269 | 11.9542 |
| NC_027318.1 | 82978132 | 119859083 | 1.4445 | 11.7309 |
| NC_027319.1 | 86795997 | 127443708 | 1.4683 | 7.8101 |
| NC_027320.1 | 58021487 | 79542378 | 1.3709 | 6.115 |
| NC_027321.1 | 63420196 | 93484531 | 1.474 | 11.4628 |
| NC_027322.1 | 49854004 | 68688932 | 1.3778 | 5.3644 |
| NC_027323.1 | 48650976 | 64992333 | 1.3359 | 10.0661 |
| NC_027324.1 | 51481326 | 73643375 | 1.4305 | 7.1473 |
| NC_027325.1 | 47900953 | 72949413 | 1.5229 | 15.3179 |
| NC_027326.1 | 43943985 | 65315682 | 1.4863 | 5.9564 |
| NC_027327.1 | 39600944 | 60187026 | 1.5198 | 21.3518 |
| NC_027328.1 | 42488238 | 57526708 | 1.3539 | 6.7771 |
| NC_001960.1 | 16665 | 9346688 | 560.8574 | 656.1734 |