Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 34,378,588 |
| Mapped reads | 34,378,588 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 34,378,588 / 100% |
| Mapped reads, first in pair | 17,189,294 / 50% |
| Mapped reads, second in pair | 17,189,294 / 50% |
| Mapped reads, both in pair | 34,378,588 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 110.57 |
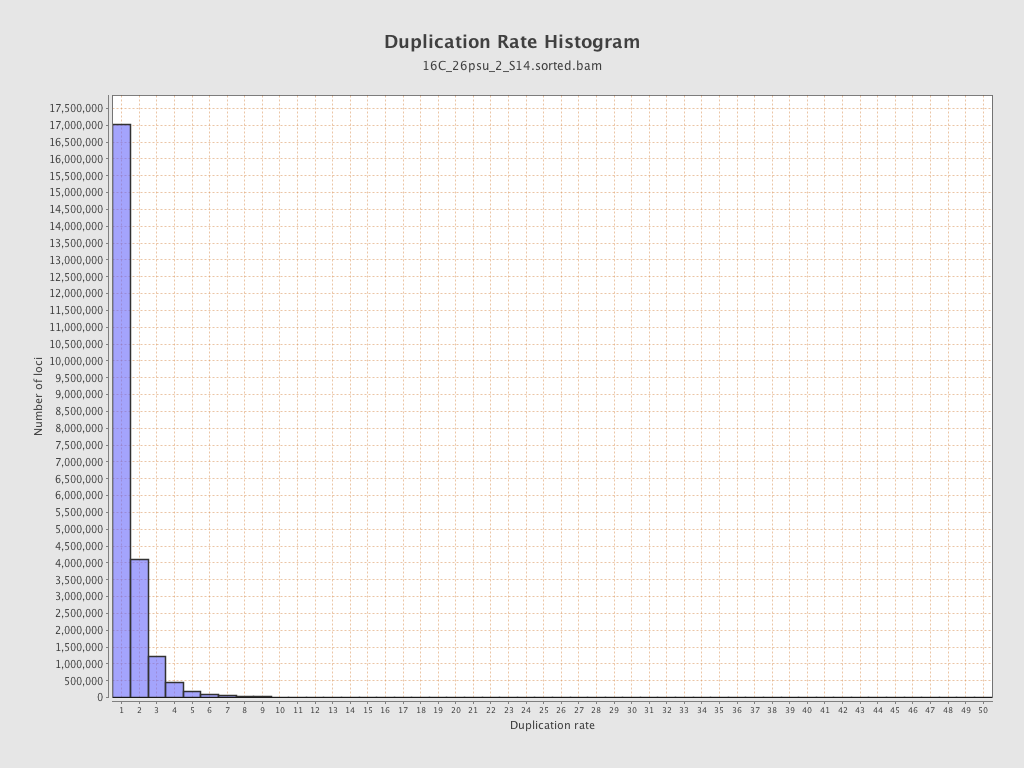
| Duplicated reads (estimated) | 11,110,088 / 32.32% |
| Duplication rate | 26.78% |
| Clipped reads | 0 / 0% |
ACGT Content
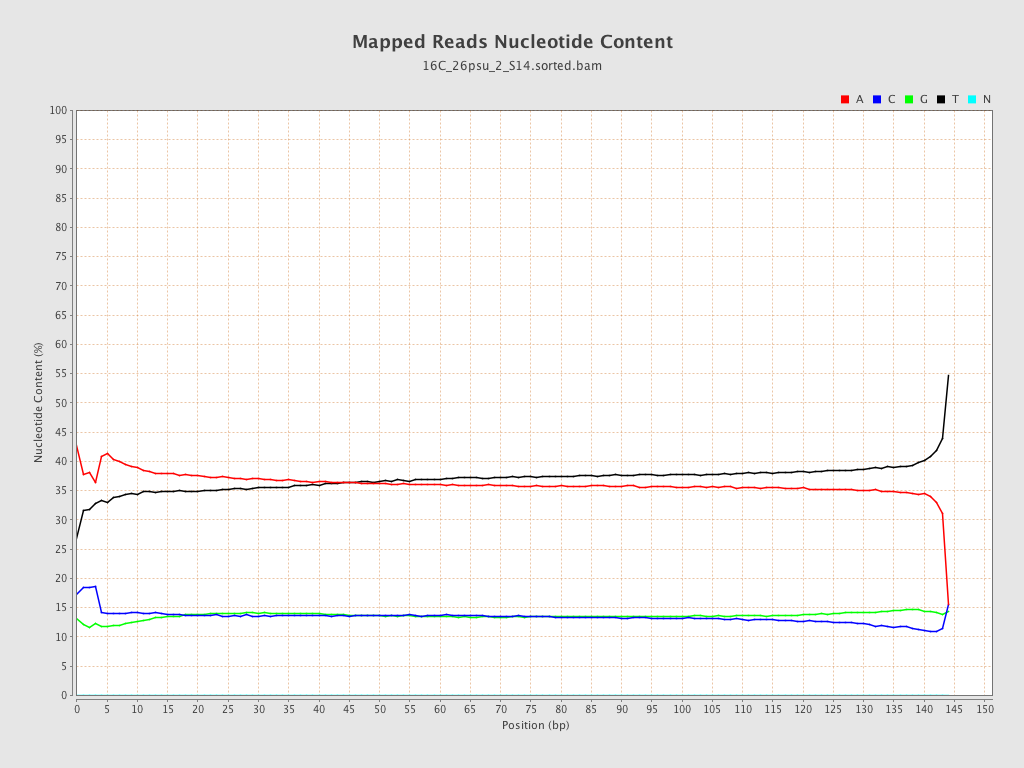
| Number/percentage of A's | 1,376,705,293 / 36.48% |
| Number/percentage of C's | 512,268,443 / 13.57% |
| Number/percentage of T's | 1,375,040,641 / 36.43% |
| Number/percentage of G's | 510,024,245 / 13.51% |
| Number/percentage of N's | 24,671 / 0% |
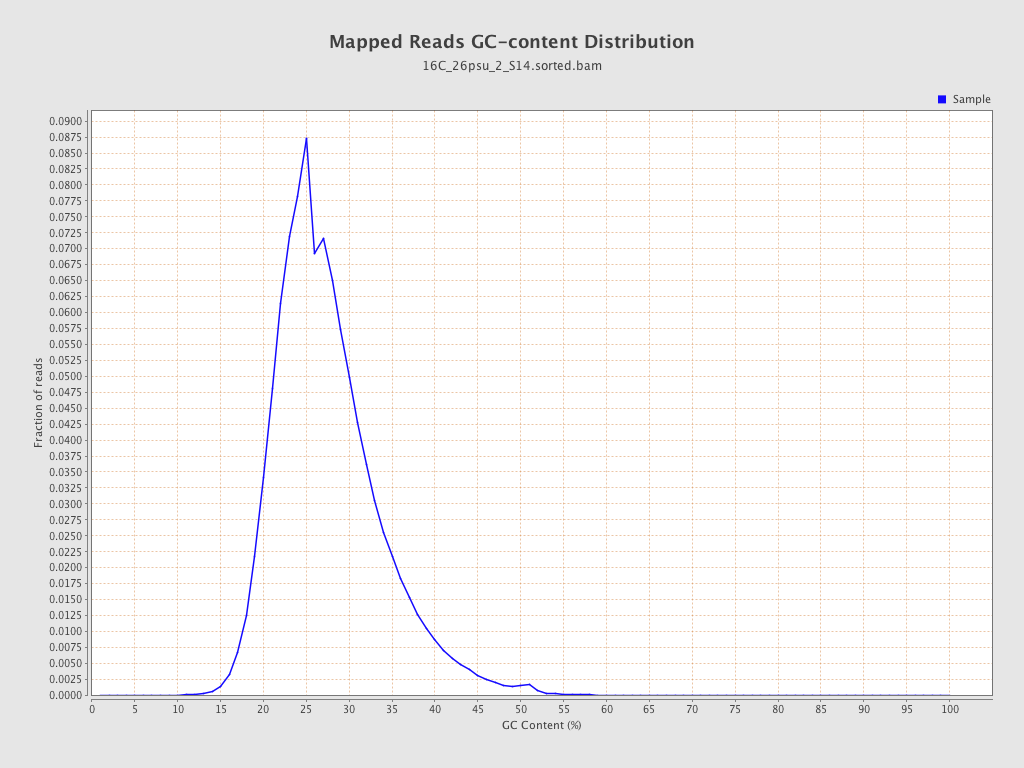
| GC Percentage | 27.09% |
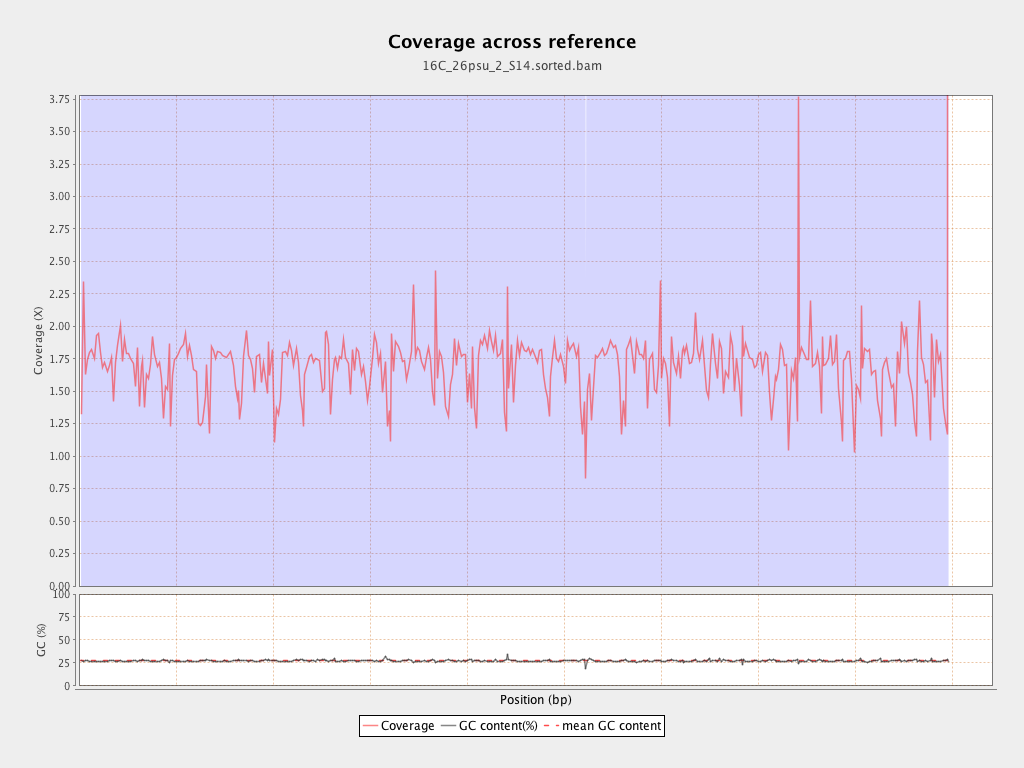
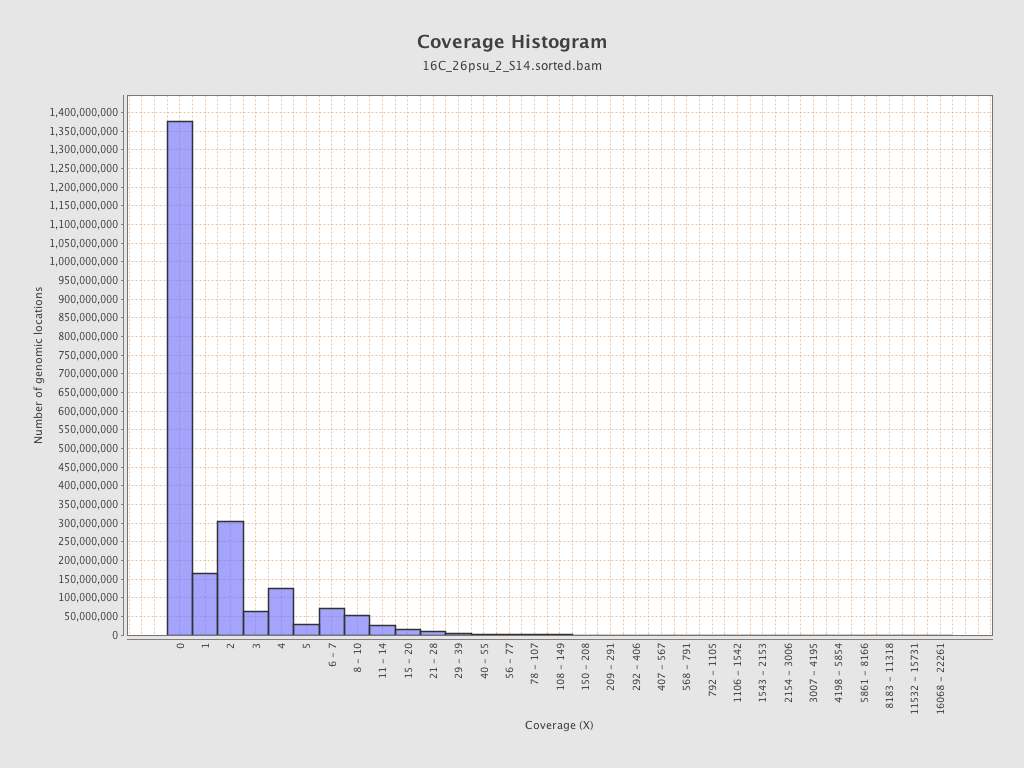
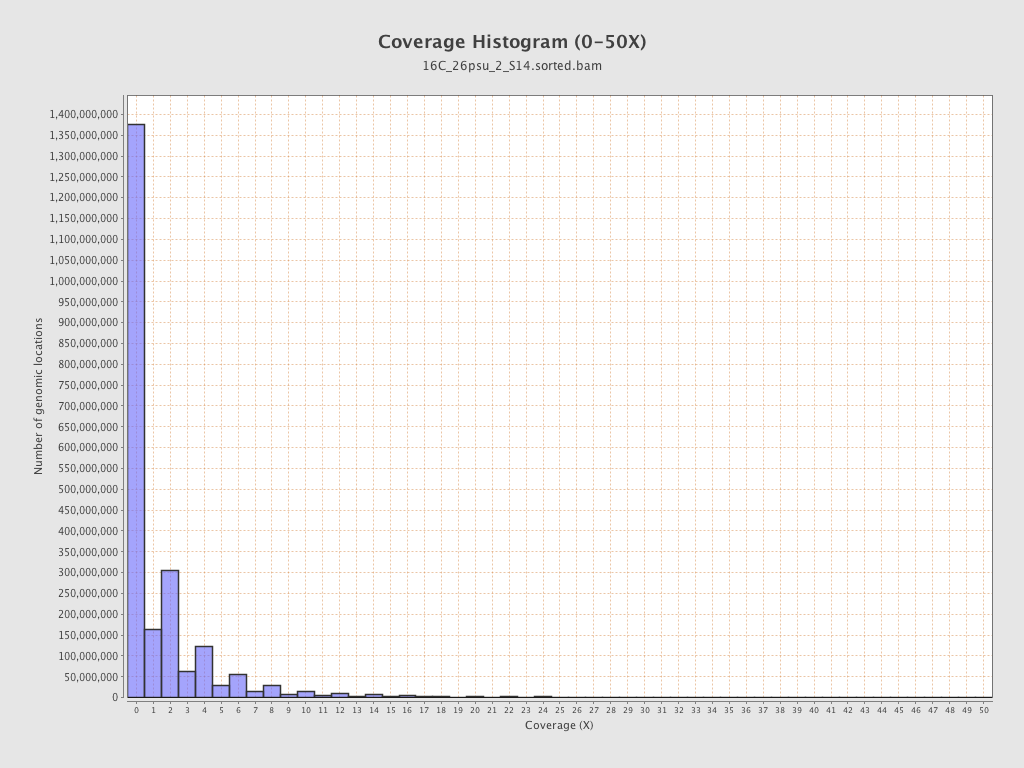
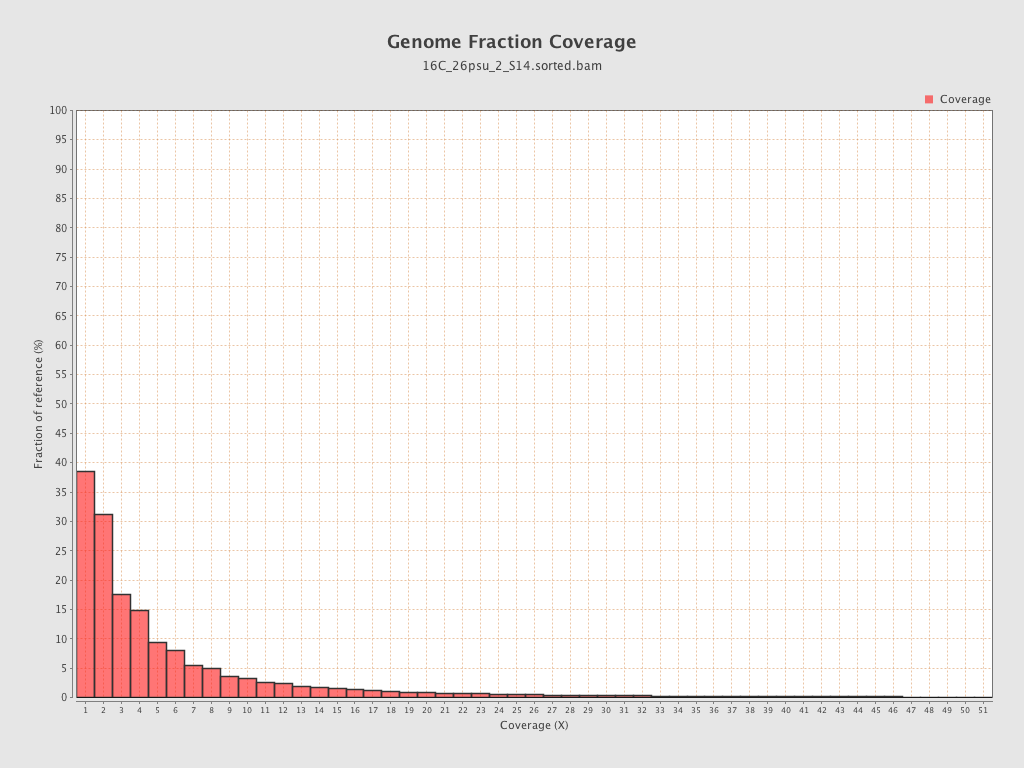
Coverage
| Mean | 1.6893 |
| Standard Deviation | 11.4299 |
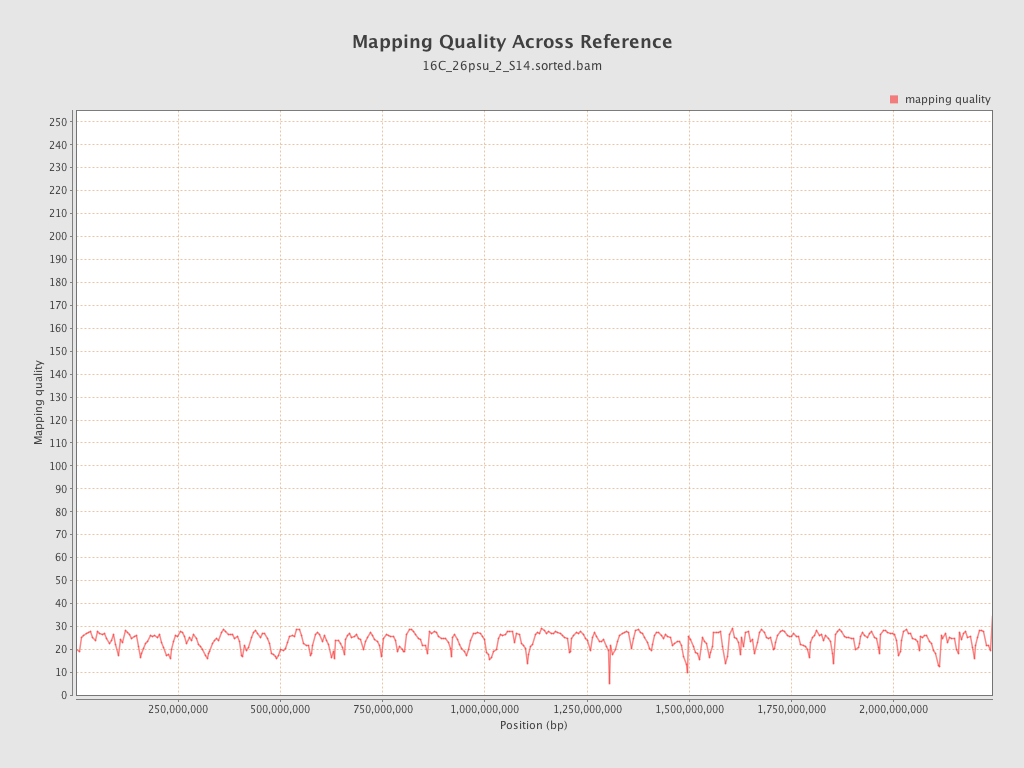
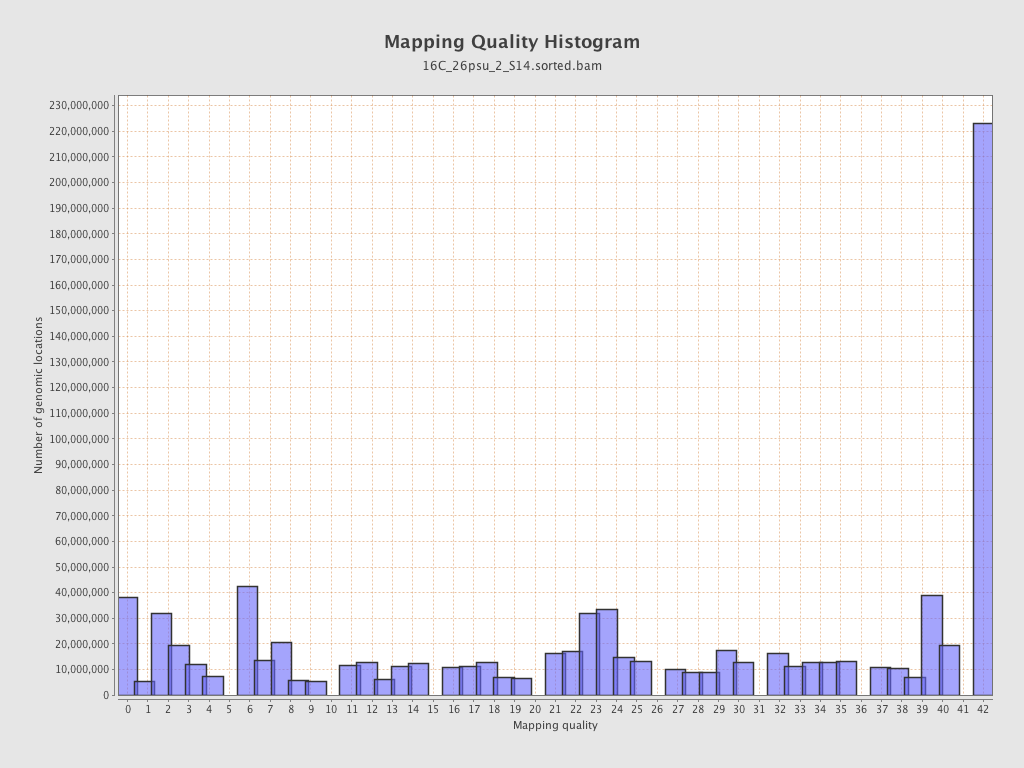
Mapping Quality
| Mean Mapping Quality | 23.68 |
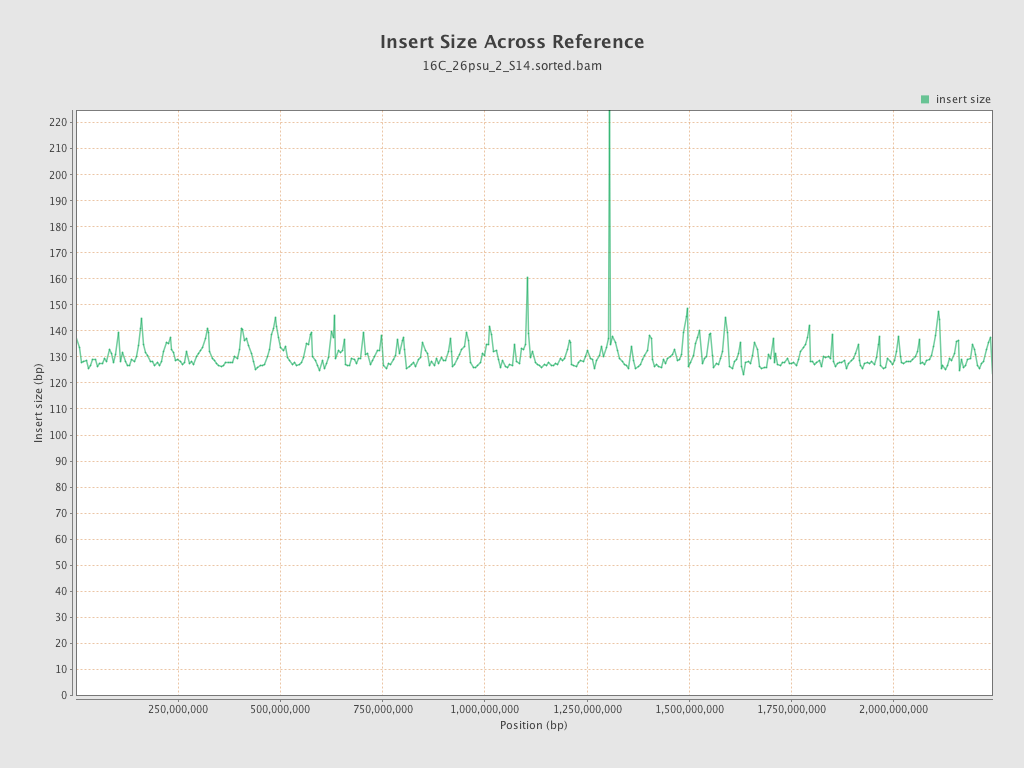
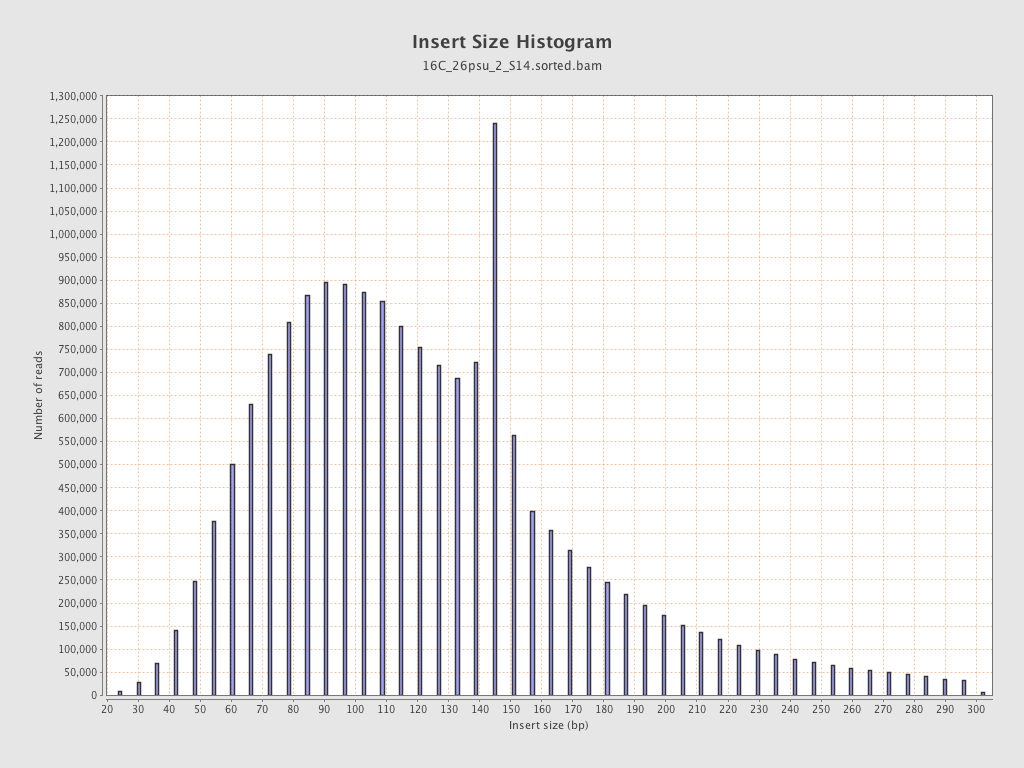
Insert size
| Mean | 130.5 |
| Standard Deviation | 60.62 |
| P25/Median/P75 | 90 / 120 / 151 |
Mismatches and indels
| General error rate | 20.85% |
| Mismatches | 760,662,059 |
| Insertions | 8,841,225 |
| Mapped reads with at least one insertion | 18.24% |
| Deletions | 6,195,636 |
| Mapped reads with at least one deletion | 15.38% |
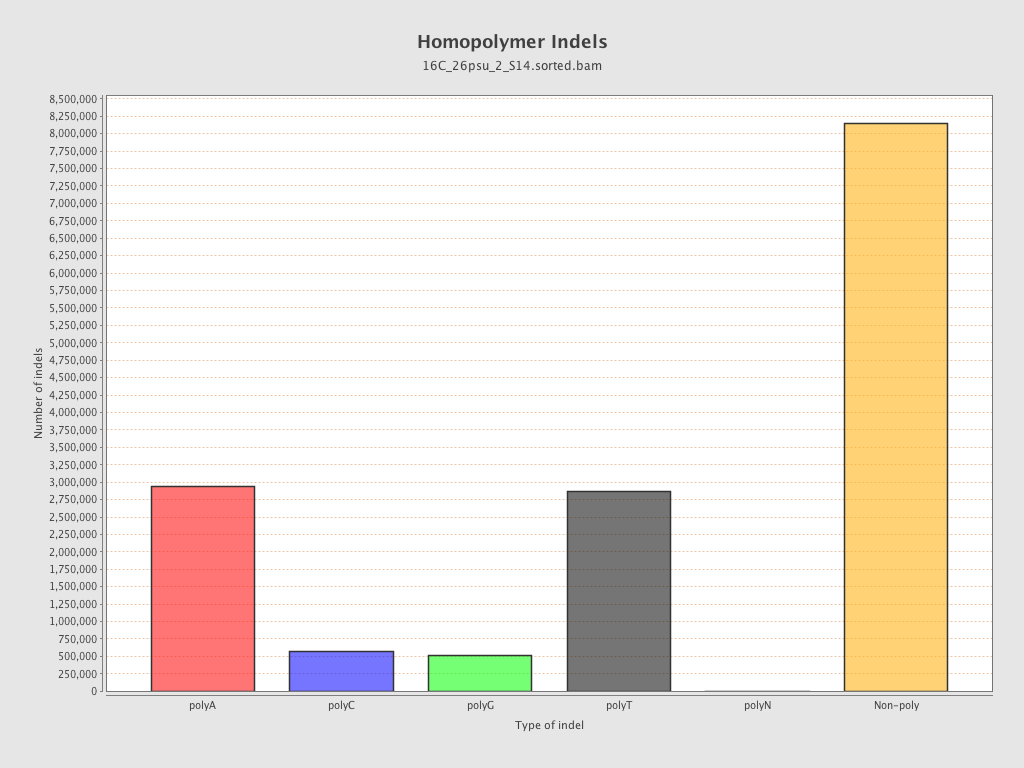
| Homopolymer indels | 45.85% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 278370823 | 1.7503 | 9.6228 |
| NC_027301.1 | 72943711 | 118756660 | 1.6281 | 6.672 |
| NC_027302.1 | 92503428 | 151406554 | 1.6368 | 5.6454 |
| NC_027303.1 | 82398023 | 140320573 | 1.703 | 7.4053 |
| NC_027304.1 | 80503876 | 132095137 | 1.6409 | 8.8091 |
| NC_027305.1 | 87043187 | 141043011 | 1.6204 | 12.0536 |
| NC_027306.1 | 58785265 | 98039709 | 1.6678 | 8.4149 |
| NC_027307.1 | 26434011 | 44628097 | 1.6883 | 12.2958 |
| NC_027308.1 | 141712163 | 237347223 | 1.6749 | 7.0432 |
| NC_027309.1 | 116138521 | 204982243 | 1.765 | 24.8709 |
| NC_027310.1 | 93888508 | 155593237 | 1.6572 | 7.7414 |
| NC_027311.1 | 91880962 | 157960219 | 1.7192 | 11.0187 |
| NC_027312.1 | 107758822 | 185769951 | 1.7239 | 7.1361 |
| NC_027313.1 | 93901823 | 159037634 | 1.6937 | 8.2256 |
| NC_027314.1 | 103963436 | 172322444 | 1.6575 | 5.6503 |
| NC_027315.1 | 87796322 | 152497340 | 1.7369 | 9.4641 |
| NC_027316.1 | 57682537 | 96057646 | 1.6653 | 8.6609 |
| NC_027317.1 | 70695409 | 124333569 | 1.7587 | 9.1702 |
| NC_027318.1 | 82978132 | 140038697 | 1.6877 | 19.5743 |
| NC_027319.1 | 86795997 | 146479691 | 1.6876 | 8.703 |
| NC_027320.1 | 58021487 | 93567303 | 1.6126 | 5.4798 |
| NC_027321.1 | 63420196 | 113241888 | 1.7856 | 20.513 |
| NC_027322.1 | 49854004 | 84227715 | 1.6895 | 7.4828 |
| NC_027323.1 | 48650976 | 76966425 | 1.582 | 6.5642 |
| NC_027324.1 | 51481326 | 85546723 | 1.6617 | 6.3925 |
| NC_027325.1 | 47900953 | 77193630 | 1.6115 | 8.5797 |
| NC_027326.1 | 43943985 | 73631909 | 1.6756 | 7.2058 |
| NC_027327.1 | 39600944 | 66812733 | 1.6872 | 12.5003 |
| NC_027328.1 | 42488238 | 65765394 | 1.5478 | 9.9437 |
| NC_001960.1 | 16665 | 10297106 | 617.8881 | 946.8495 |