Summary
Globals
| Reference size | 2,240,221,656 |
| Number of reads | 43,151,604 |
| Mapped reads | 43,151,604 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 43,151,604 / 100% |
| Mapped reads, first in pair | 21,575,802 / 50% |
| Mapped reads, second in pair | 21,575,802 / 50% |
| Mapped reads, both in pair | 43,151,604 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Read min/max/mean length | 20 / 145 / 108.76 |
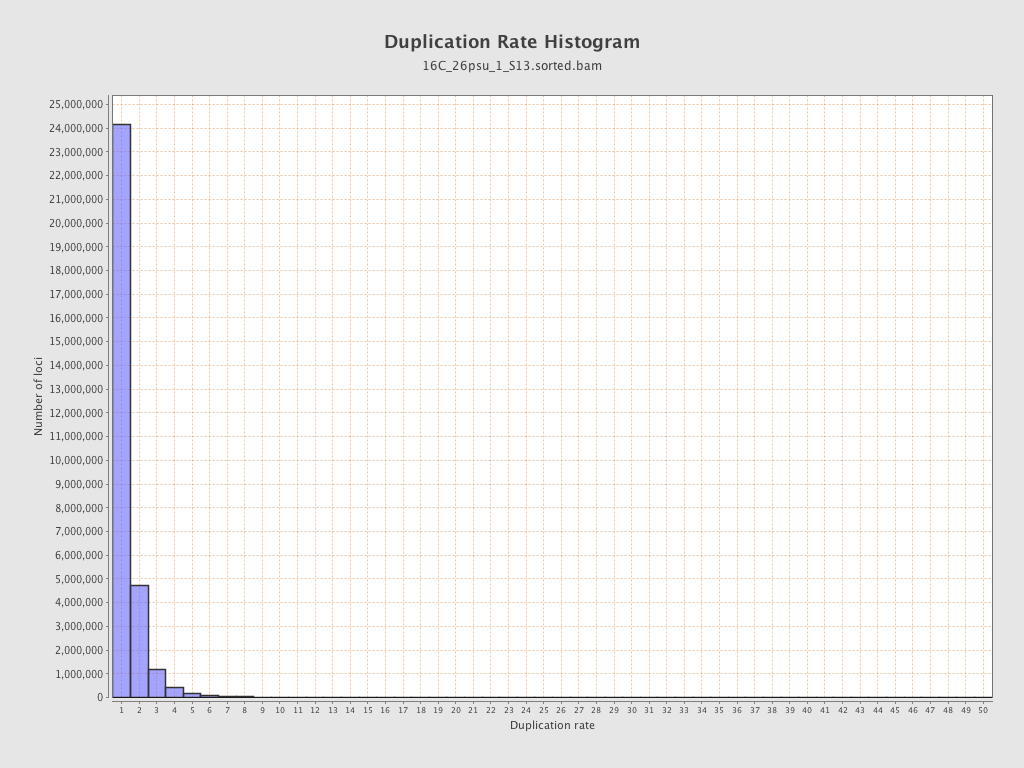
| Duplicated reads (estimated) | 12,161,108 / 28.18% |
| Duplication rate | 21.99% |
| Clipped reads | 0 / 0% |
ACGT Content
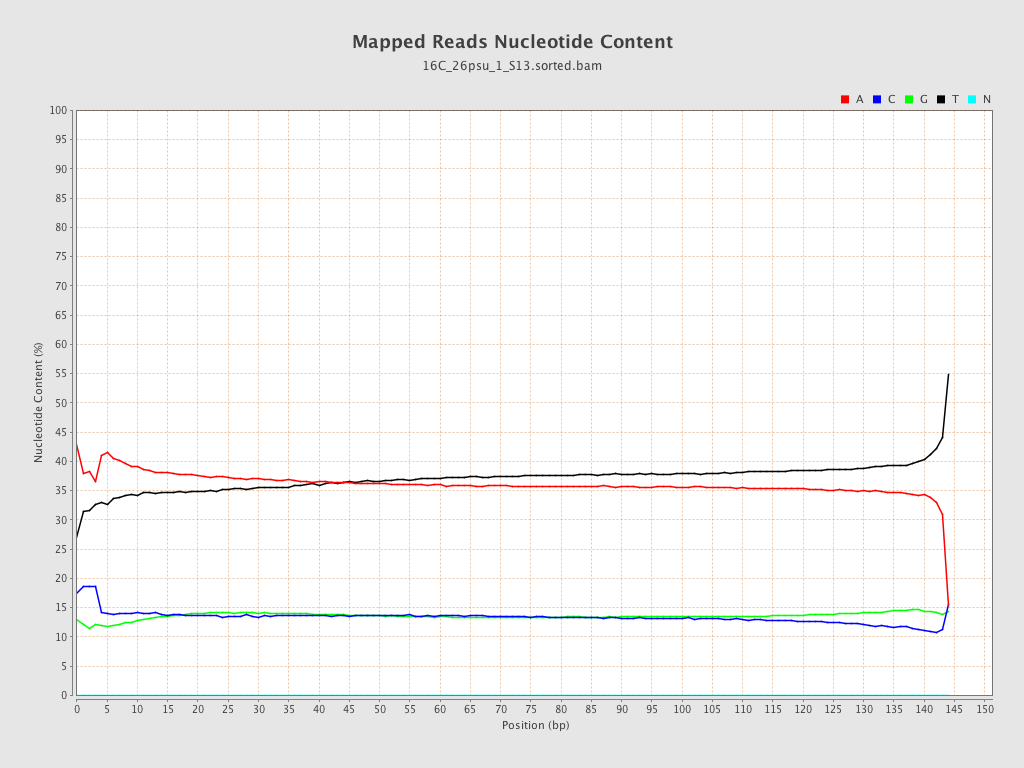
| Number/percentage of A's | 1,700,108,382 / 36.49% |
| Number/percentage of C's | 631,343,923 / 13.55% |
| Number/percentage of T's | 1,699,080,232 / 36.47% |
| Number/percentage of G's | 628,275,602 / 13.49% |
| Number/percentage of N's | 31,127 / 0% |
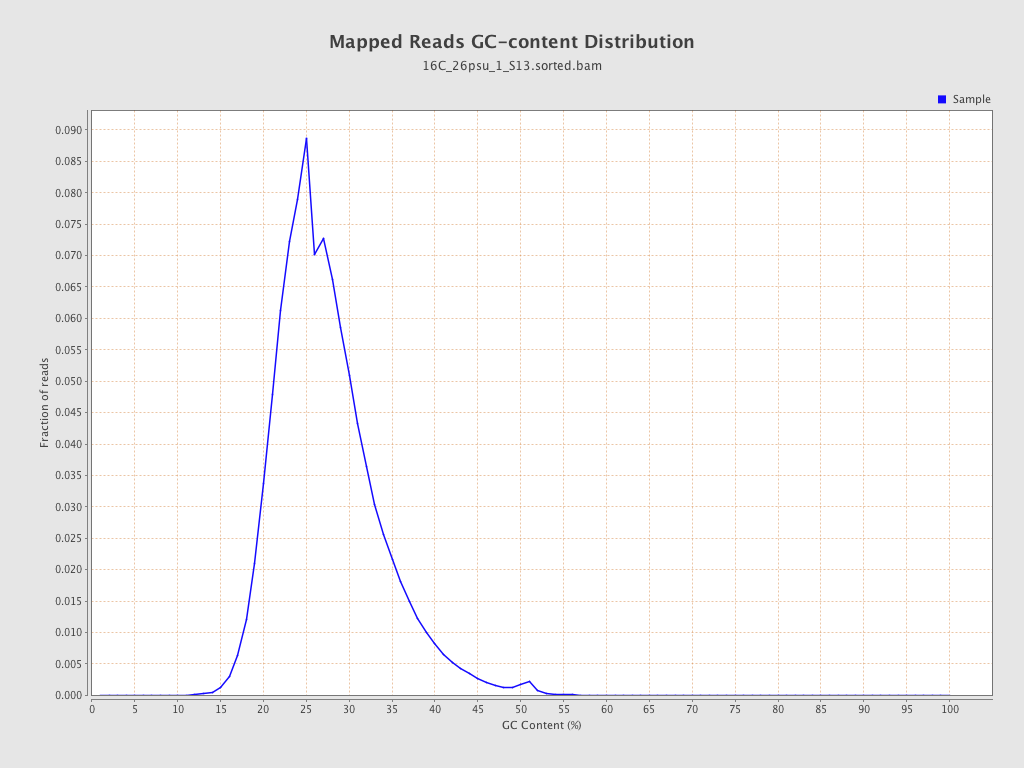
| GC Percentage | 27.04% |
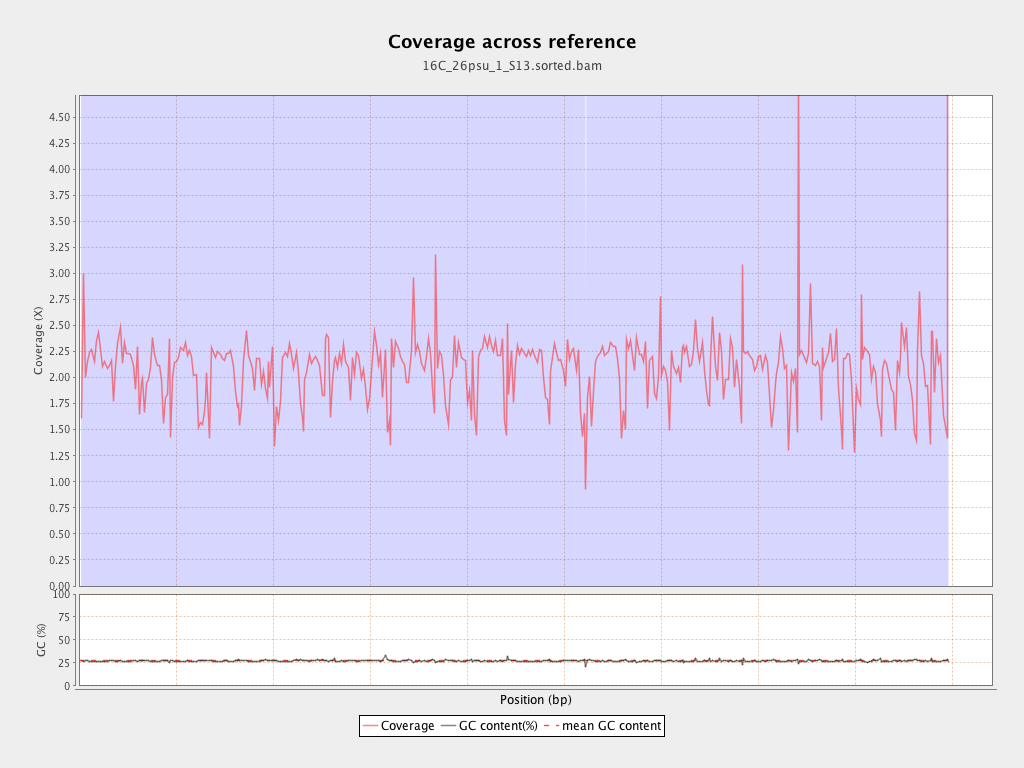
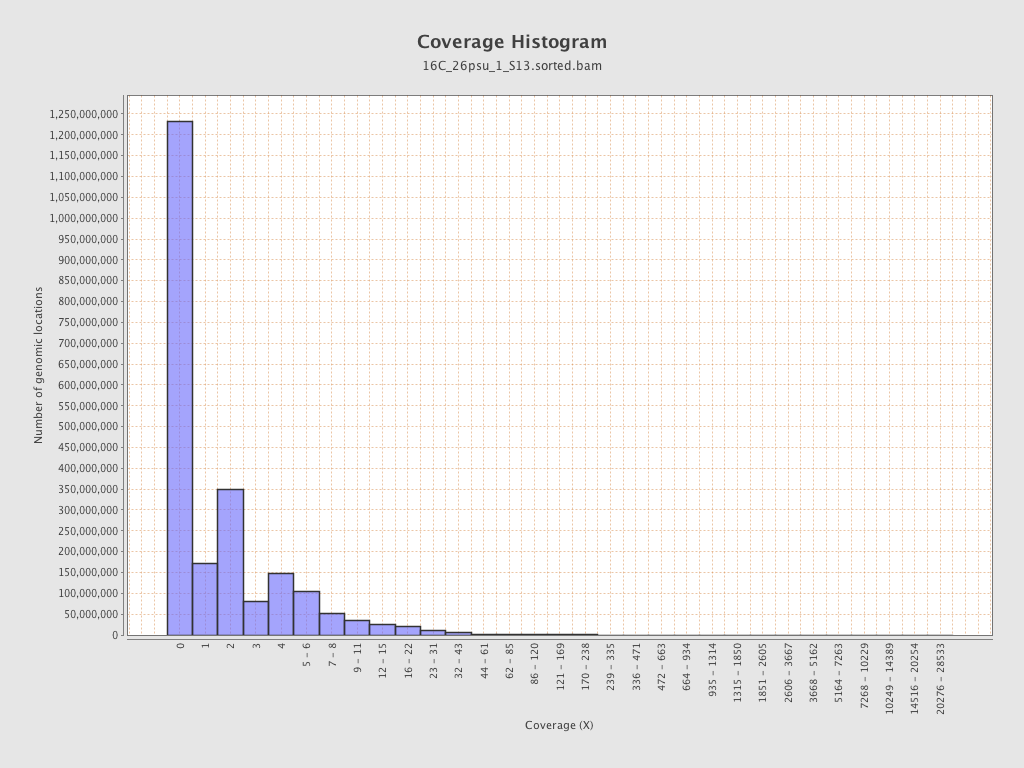
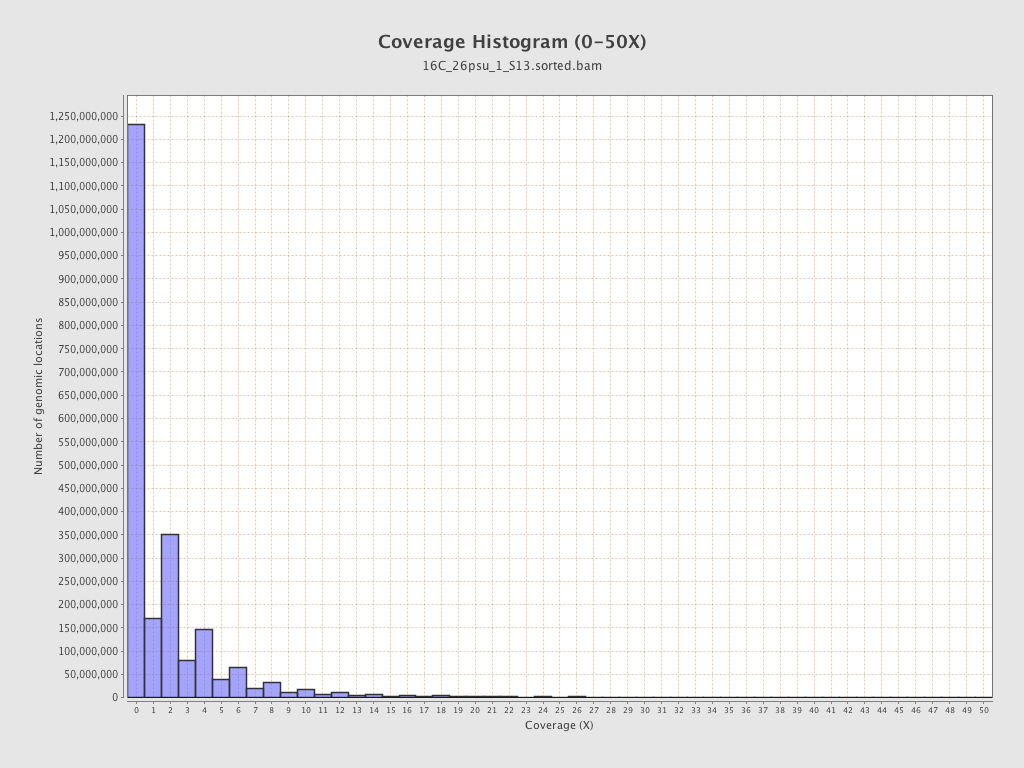
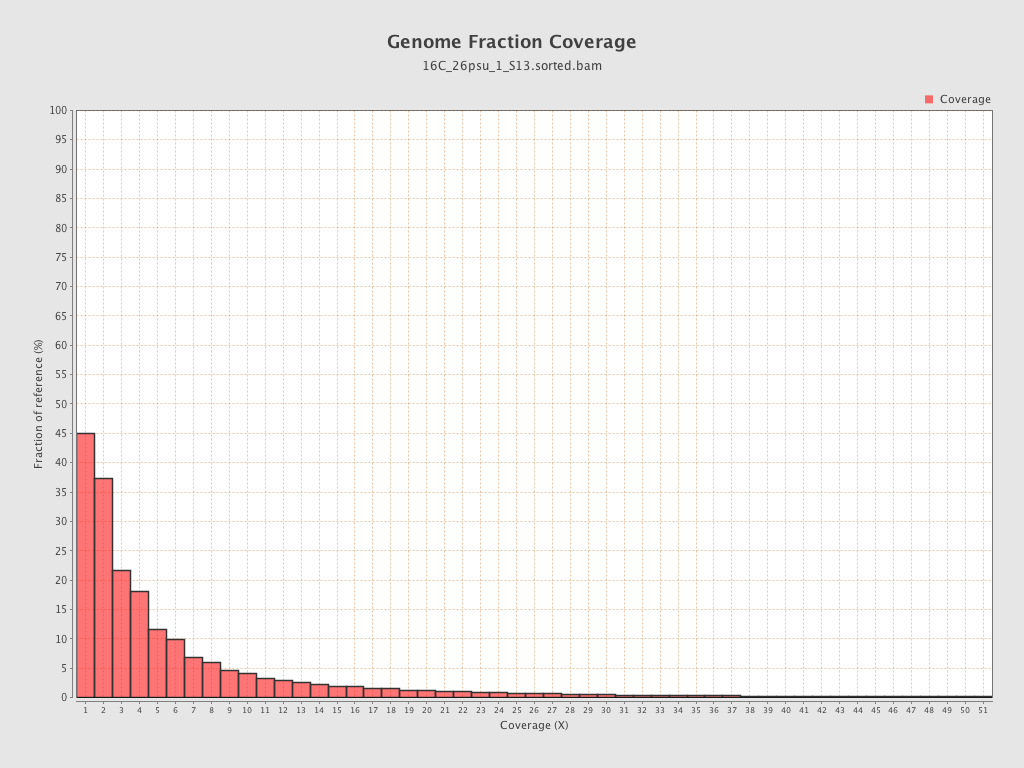
Coverage
| Mean | 2.0853 |
| Standard Deviation | 15.4161 |
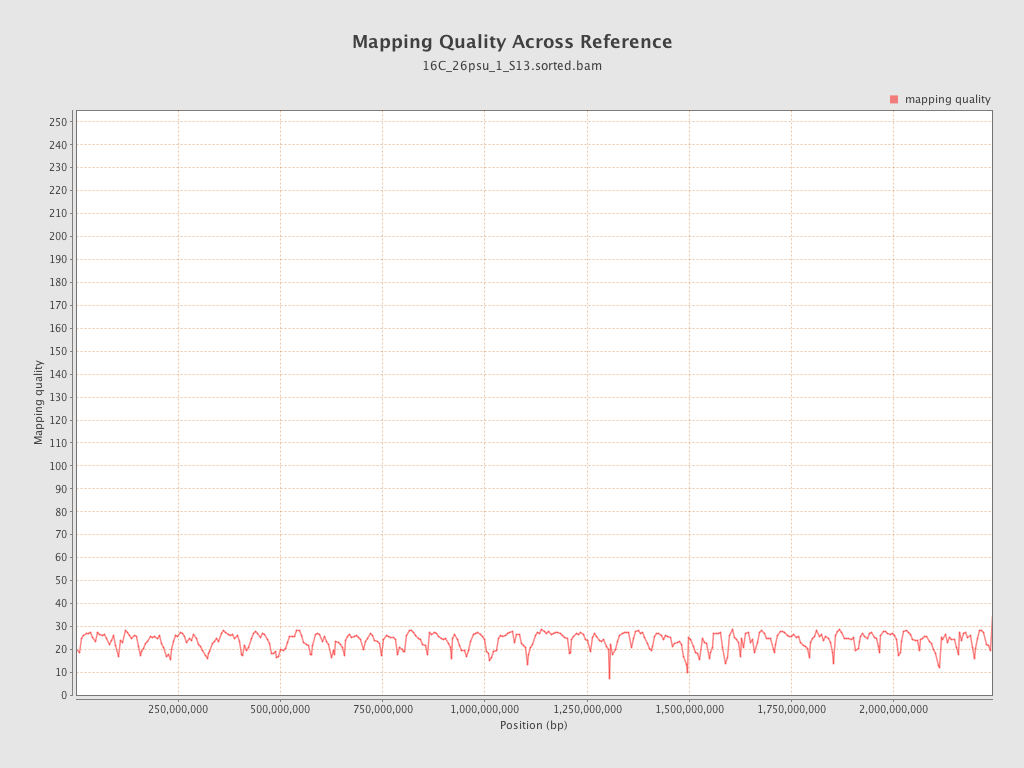
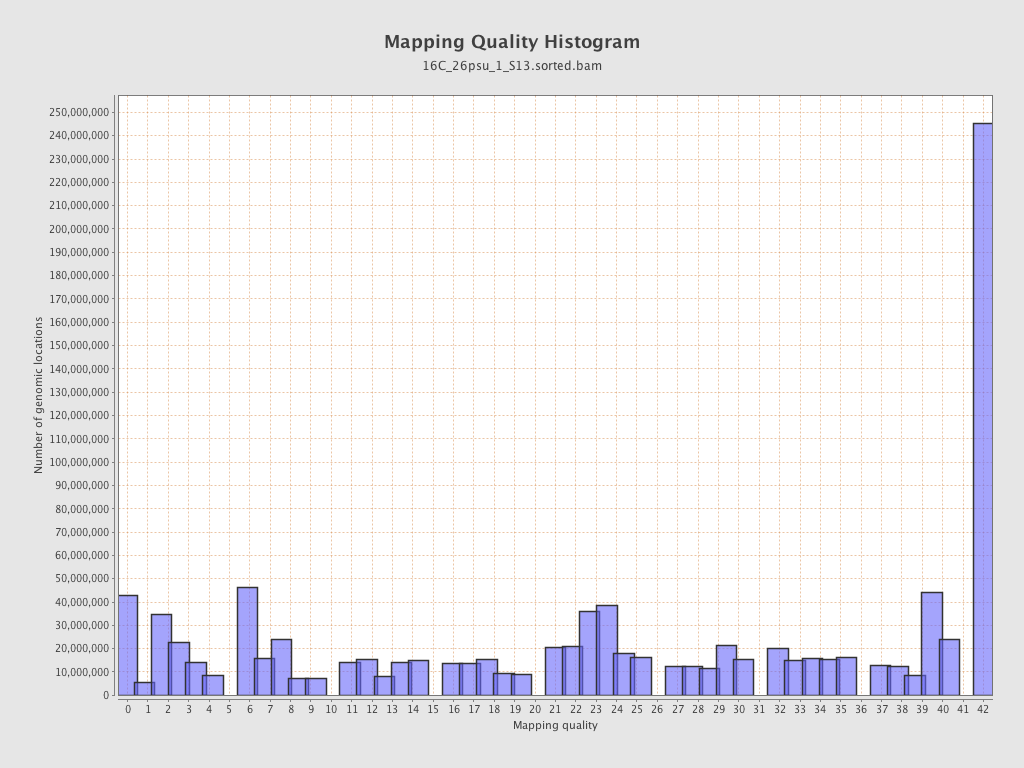
Mapping Quality
| Mean Mapping Quality | 23.53 |
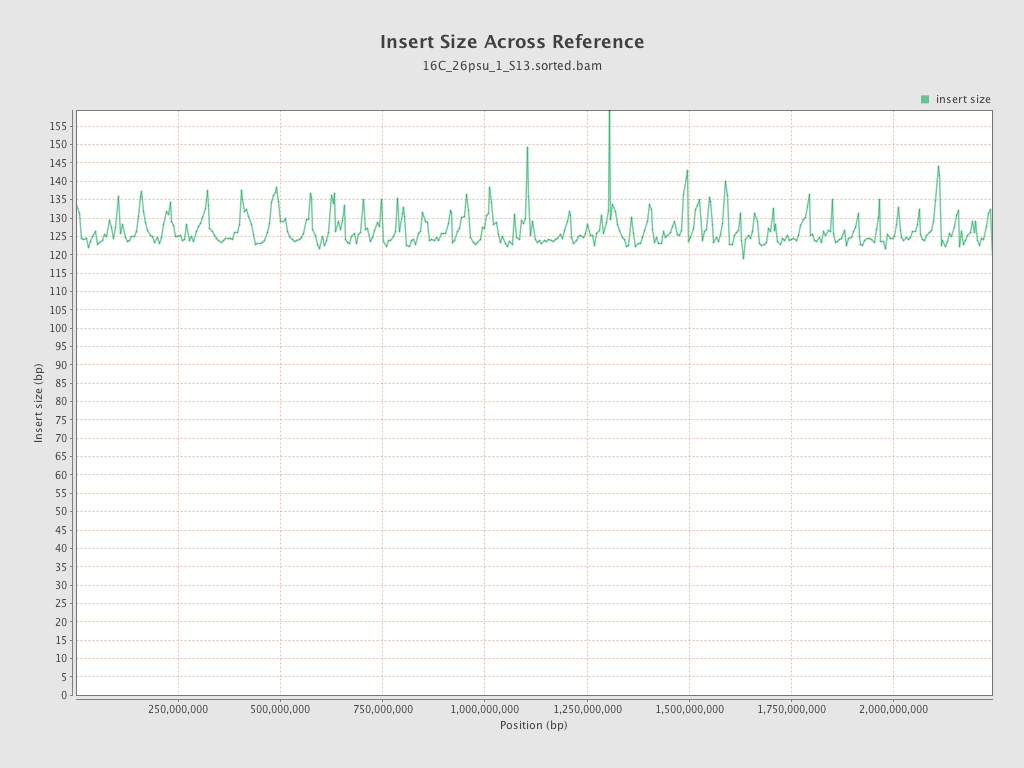
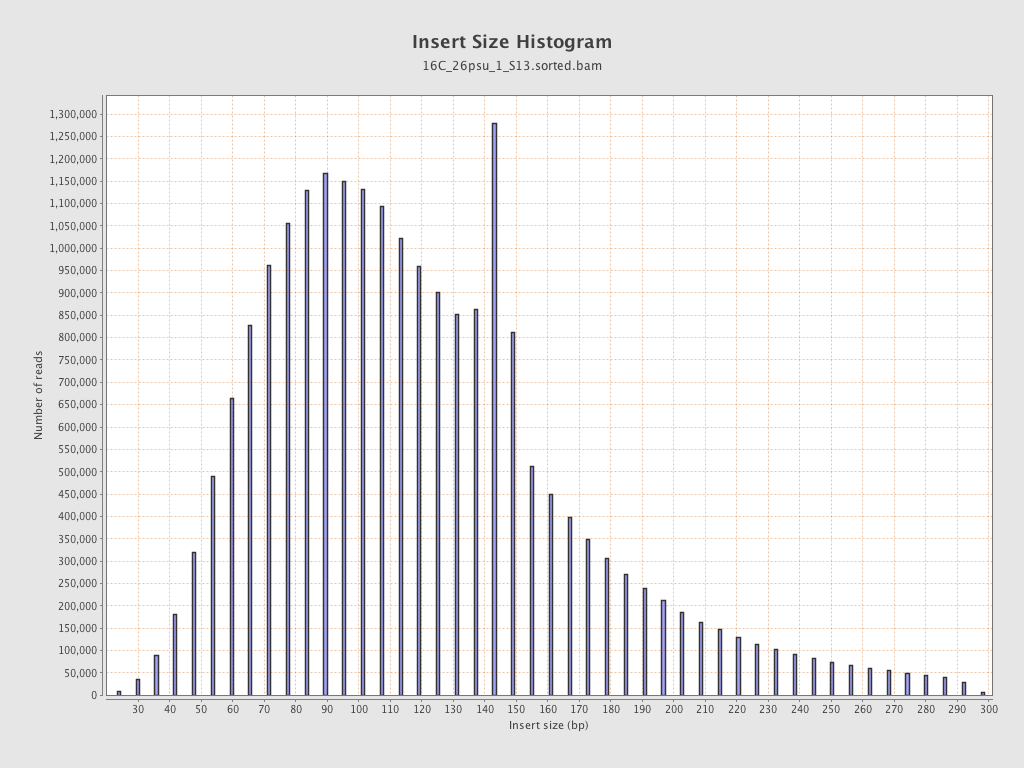
Insert size
| Mean | 126.82 |
| Standard Deviation | 58.28 |
| P25/Median/P75 | 88 / 116 / 149 |
Mismatches and indels
| General error rate | 21.16% |
| Mismatches | 952,291,310 |
| Insertions | 11,390,891 |
| Mapped reads with at least one insertion | 18.6% |
| Deletions | 7,804,249 |
| Mapped reads with at least one deletion | 15.41% |
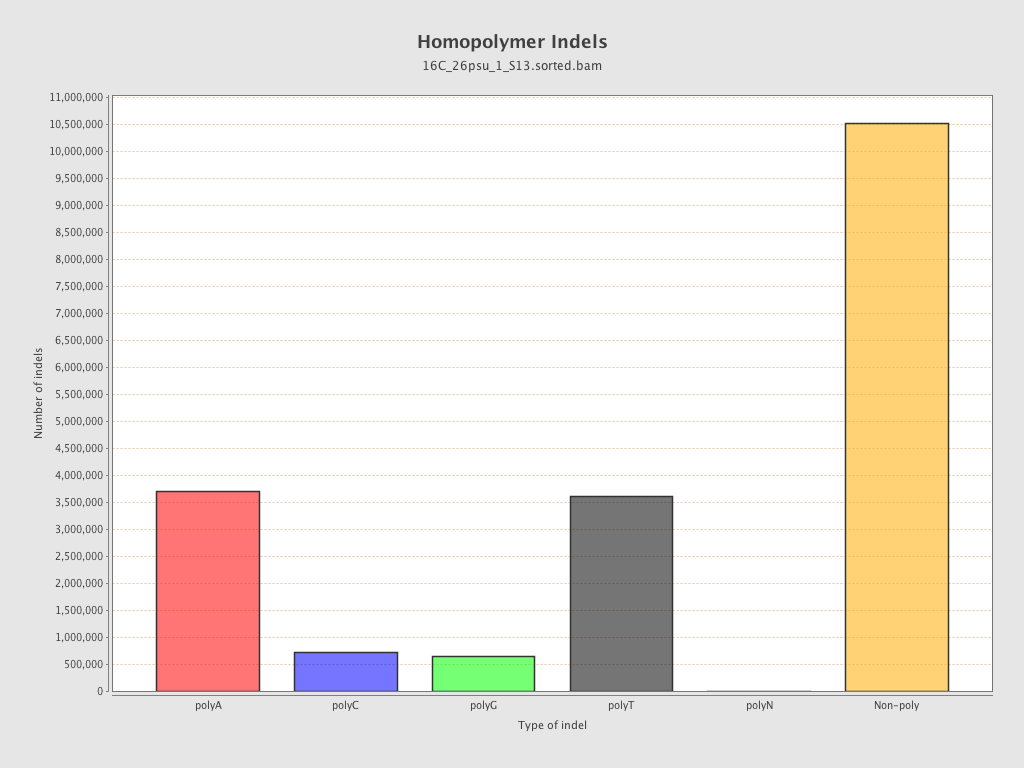
| Homopolymer indels | 45.22% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NC_027300.1 | 159038749 | 345557754 | 2.1728 | 13.233 |
| NC_027301.1 | 72943711 | 145063271 | 1.9887 | 8.0613 |
| NC_027302.1 | 92503428 | 185545311 | 2.0058 | 6.5835 |
| NC_027303.1 | 82398023 | 173148172 | 2.1014 | 10.4099 |
| NC_027304.1 | 80503876 | 162051440 | 2.013 | 10.8324 |
| NC_027305.1 | 87043187 | 172439143 | 1.9811 | 16.1485 |
| NC_027306.1 | 58785265 | 120347279 | 2.0472 | 10.477 |
| NC_027307.1 | 26434011 | 55550751 | 2.1015 | 20.474 |
| NC_027308.1 | 141712163 | 291028877 | 2.0537 | 10.549 |
| NC_027309.1 | 116138521 | 255270729 | 2.198 | 31.7617 |
| NC_027310.1 | 93888508 | 192252615 | 2.0477 | 9.0258 |
| NC_027311.1 | 91880962 | 193593164 | 2.107 | 13.0932 |
| NC_027312.1 | 107758822 | 228745300 | 2.1228 | 9.4086 |
| NC_027313.1 | 93901823 | 195068025 | 2.0774 | 11.0445 |
| NC_027314.1 | 103963436 | 212010035 | 2.0393 | 6.7599 |
| NC_027315.1 | 87796322 | 188620223 | 2.1484 | 11.9049 |
| NC_027316.1 | 57682537 | 116689703 | 2.023 | 10.8364 |
| NC_027317.1 | 70695409 | 153458485 | 2.1707 | 12.8544 |
| NC_027318.1 | 82978132 | 174299540 | 2.1005 | 28.5264 |
| NC_027319.1 | 86795997 | 180213078 | 2.0763 | 10.281 |
| NC_027320.1 | 58021487 | 116040481 | 2 | 5.964 |
| NC_027321.1 | 63420196 | 141423755 | 2.2299 | 29.4096 |
| NC_027322.1 | 49854004 | 105610619 | 2.1184 | 9.7755 |
| NC_027323.1 | 48650976 | 94735215 | 1.9472 | 7.8011 |
| NC_027324.1 | 51481326 | 106433962 | 2.0674 | 9.6266 |
| NC_027325.1 | 47900953 | 94641048 | 1.9758 | 9.8126 |
| NC_027326.1 | 43943985 | 90053420 | 2.0493 | 9.2153 |
| NC_027327.1 | 39600944 | 83249681 | 2.1022 | 15.9418 |
| NC_027328.1 | 42488238 | 81841805 | 1.9262 | 14.8924 |
| NC_001960.1 | 16665 | 16663143 | 999.8886 | 1,423.305 |