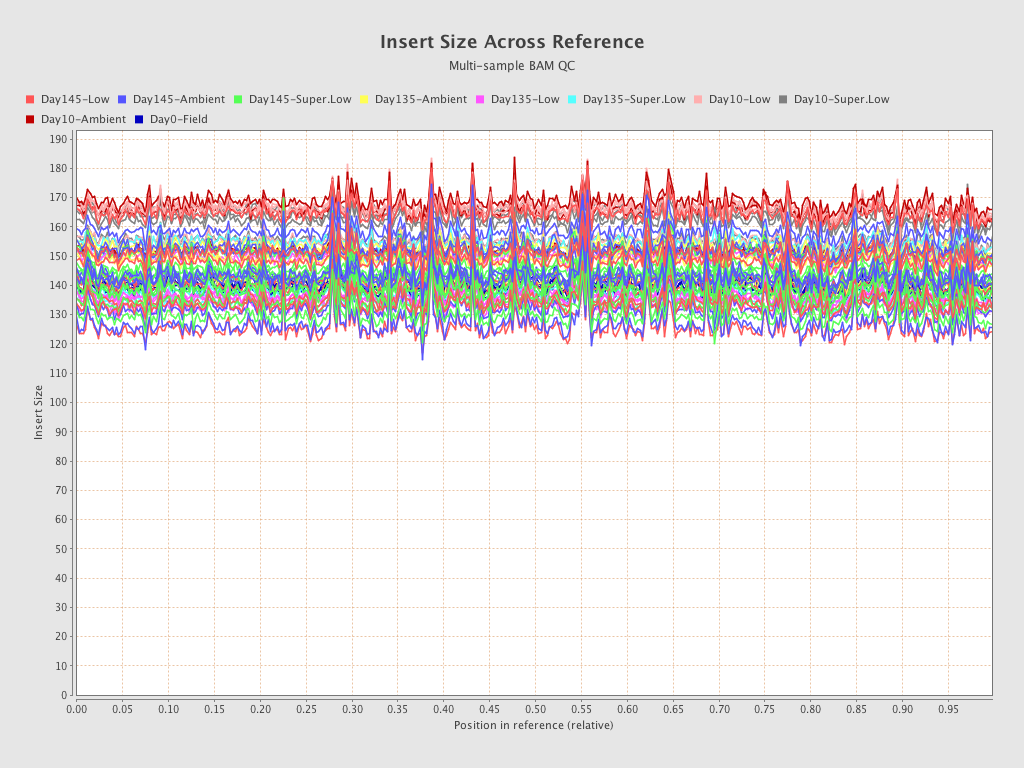
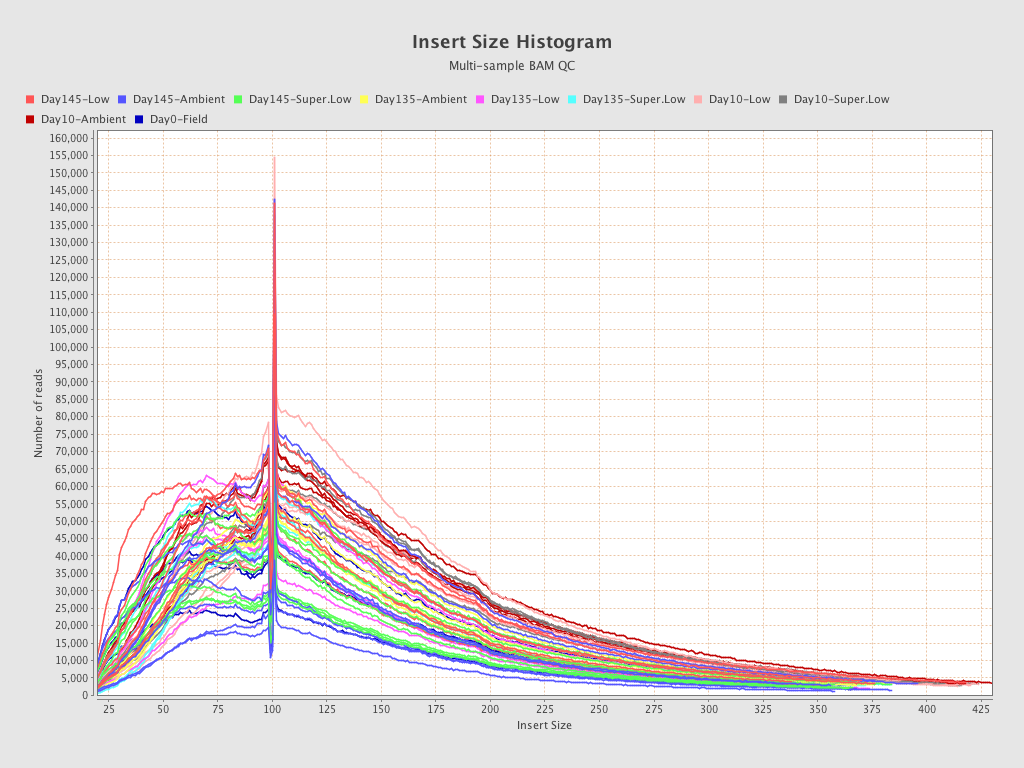
| EPI-168 (Day135-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-168_S11_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-136 (Day10-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-136_S36_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-182 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-182_S17_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-42 (Day0-Field) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-42_S39_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-220 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-181 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-181_S16_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-187 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-187_S20_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-185 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-185_S19_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-127 (Day10-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-127_S33_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-104 (Day10-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-104_S28_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-176 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-176_S15_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-153 (Day135-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-153_S4_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-152 (Day135-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-152_S3_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-209 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-209_S29_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-208 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-208_S28_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-175 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-175_S14_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-44 (Day0-Field) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-44_S41_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-226 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-128 (Day10-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-128_S34_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-199 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-199_S24_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-230 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-230_S37_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-205 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-103 (Day10-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-103_S27_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-120 (Day10-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-120_S32_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-135 (Day10-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-135_S35_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-159 (Day135-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-159_S6_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-167 (Day135-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-167_S10_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-145 (Day10-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-145_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-169 (Day135-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-169_S12_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-200 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-200_S25_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-41 (Day0-Field) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-41_S38_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-43 (Day0-Field) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-43_S40_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-111 (Day10-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-111_S29_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-221 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-188 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-188_S21_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-119 (Day10-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-119_S31_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-194 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-194_S23_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-162 (Day135-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-162_S9_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-161 (Day135-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-161_S8_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-193 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-193_S22_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-214 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-184 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-003/EPI-184_S18_L003_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-215 (Day145-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-227 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-229 (Day145-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-229_S36_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-151 (Day135-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-151_S2_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-113 (Day10-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-113_S30_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-154 (Day135-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-154_S5_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-206 (Day145-Ambient) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807-004/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-160 (Day135-Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-160_S7_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-170 (Day135-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0807/EPI-170_S13_L002_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |
| EPI-143 (Day10-Super.Low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190822/dedup_bams/0809-005/EPI-143_S37_L005_R1_001_val_1_bismark_bt2_pe.deduplicated.sorted_stats |

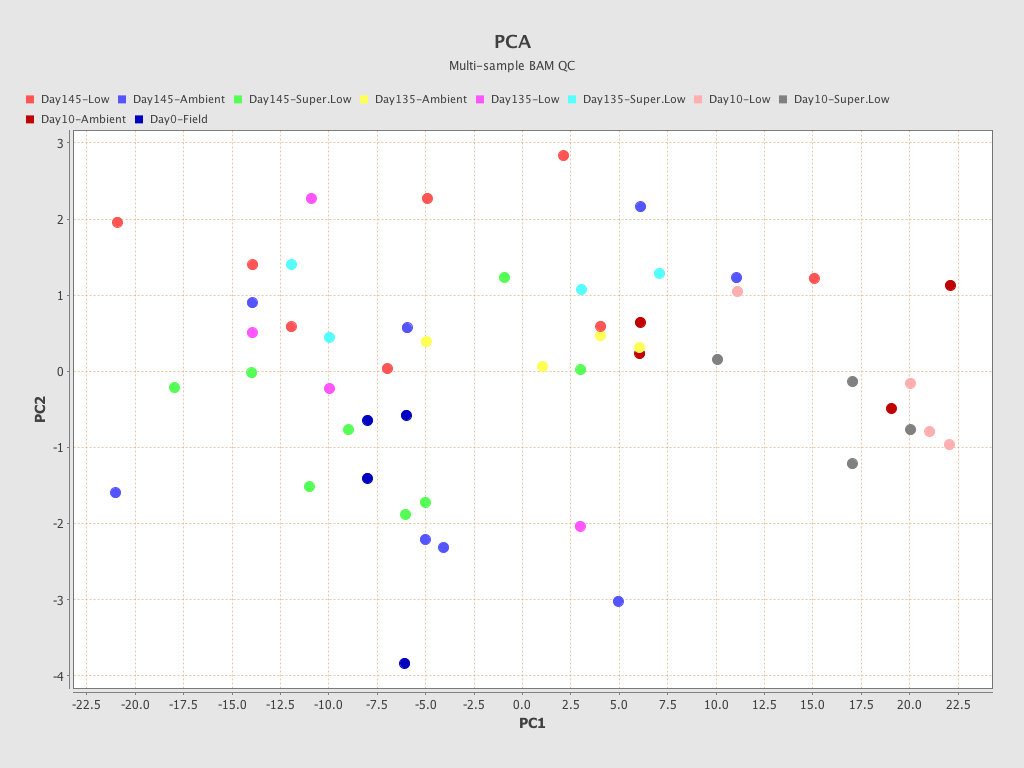
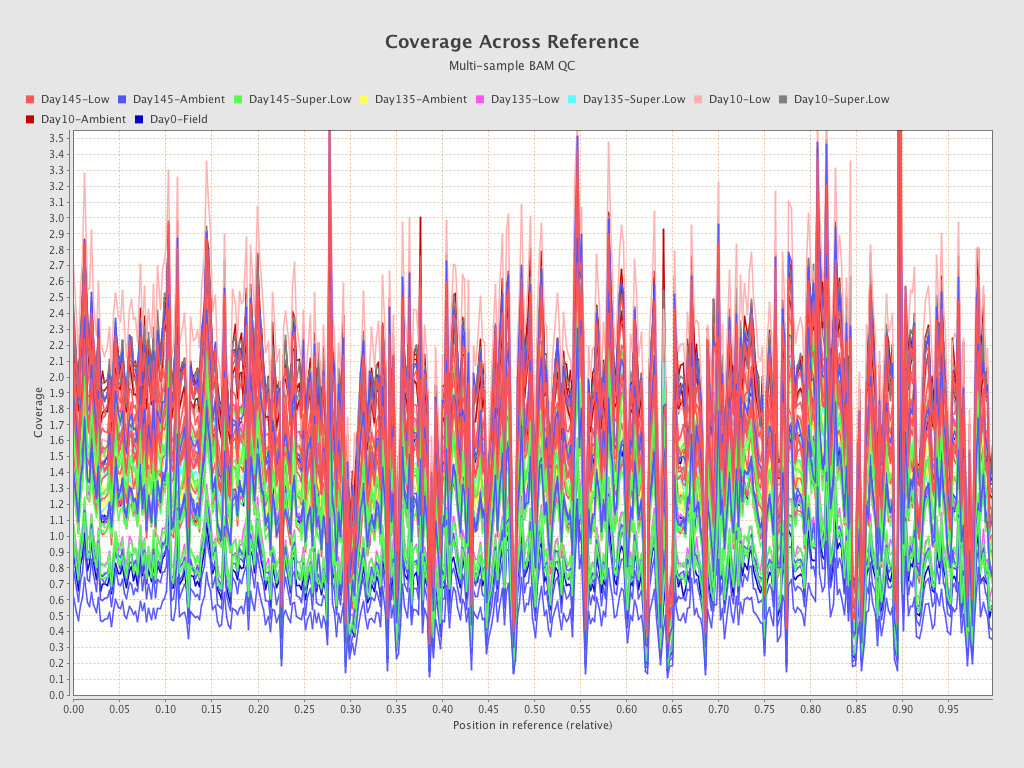
.png)