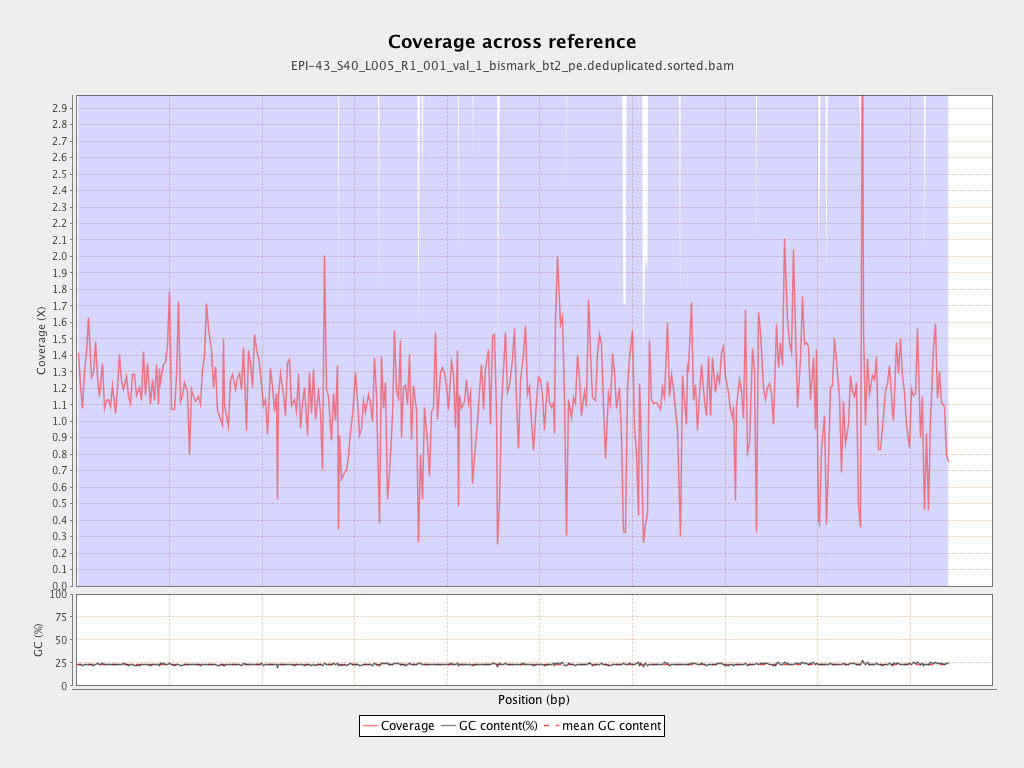
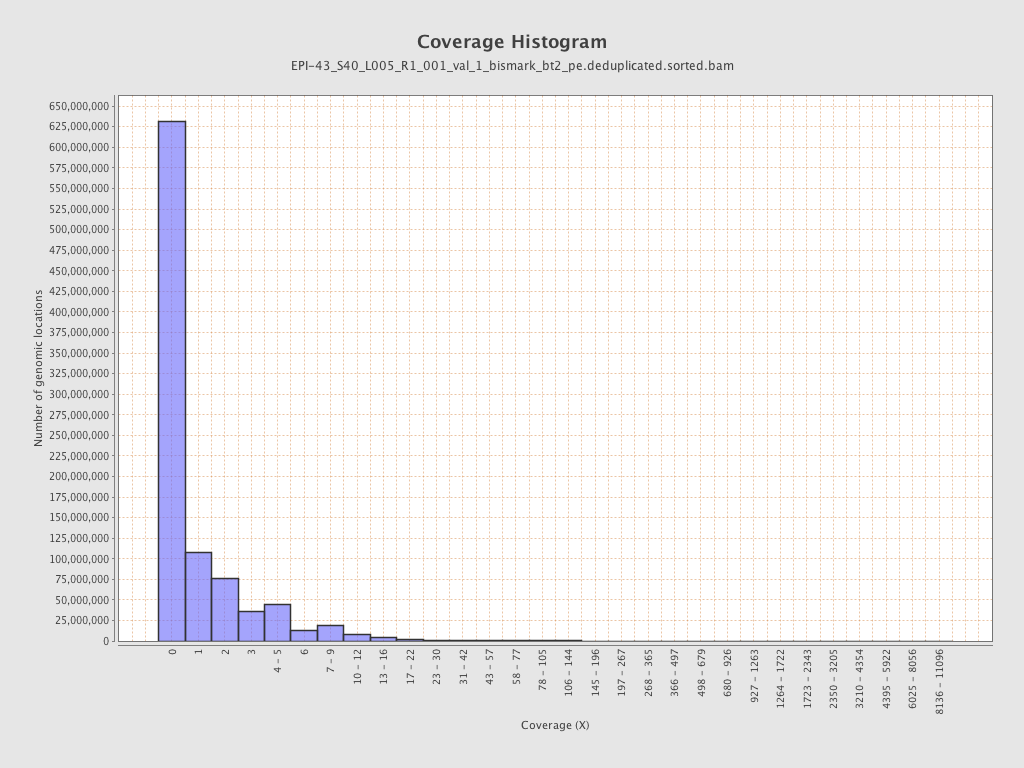
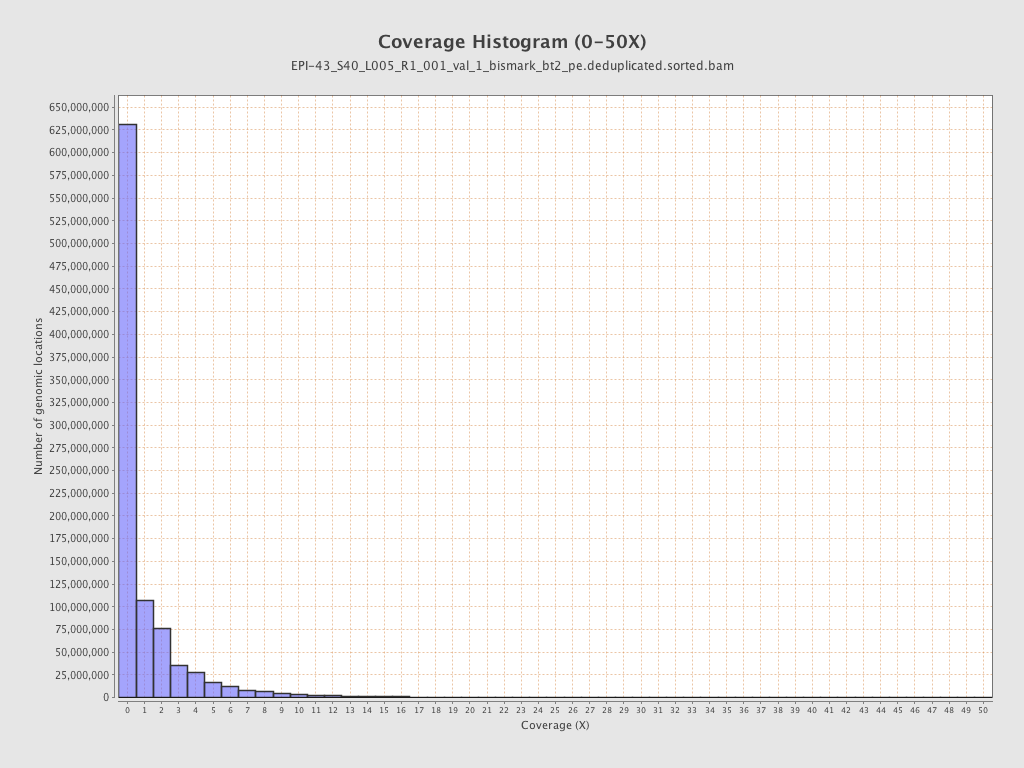
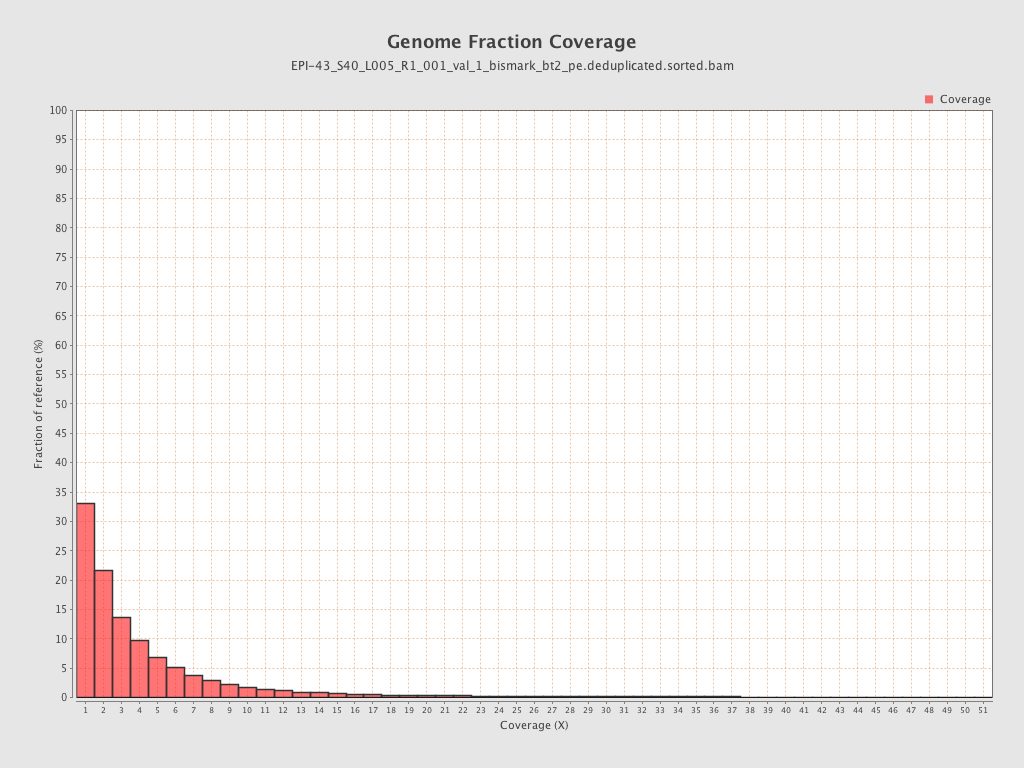
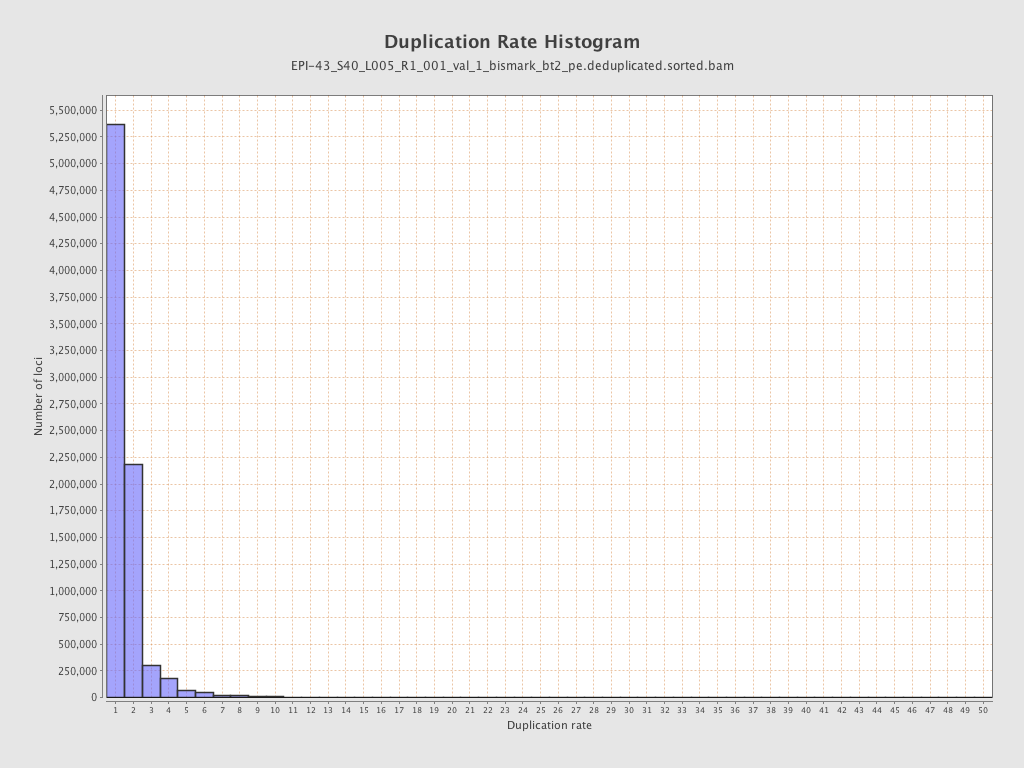
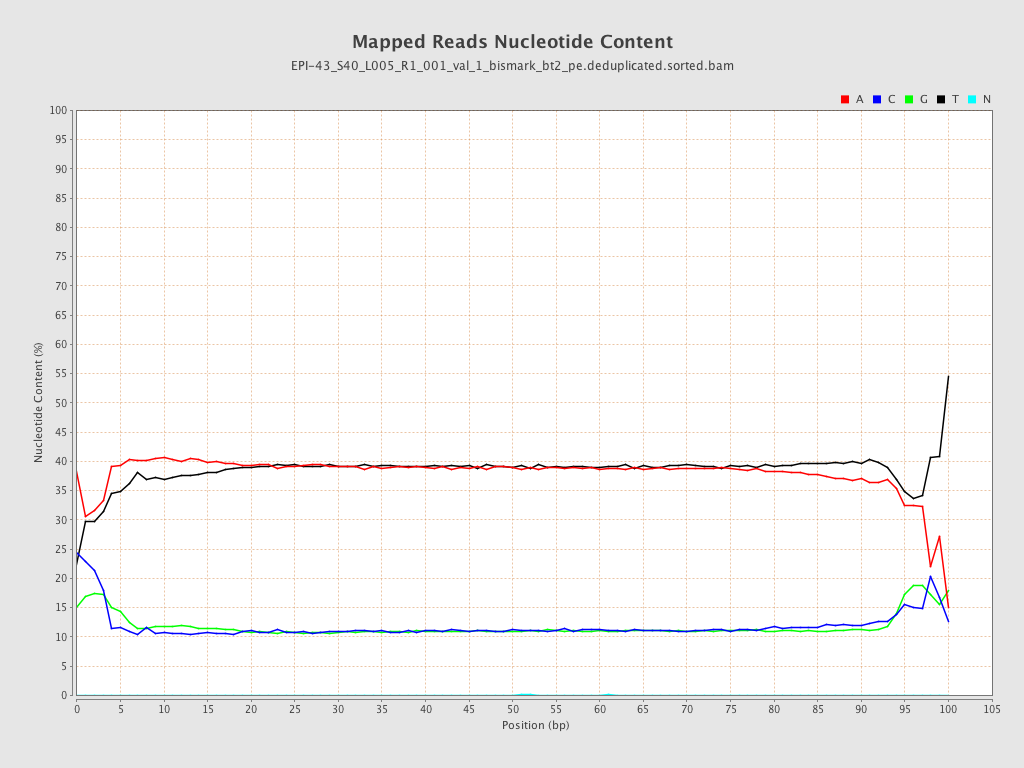
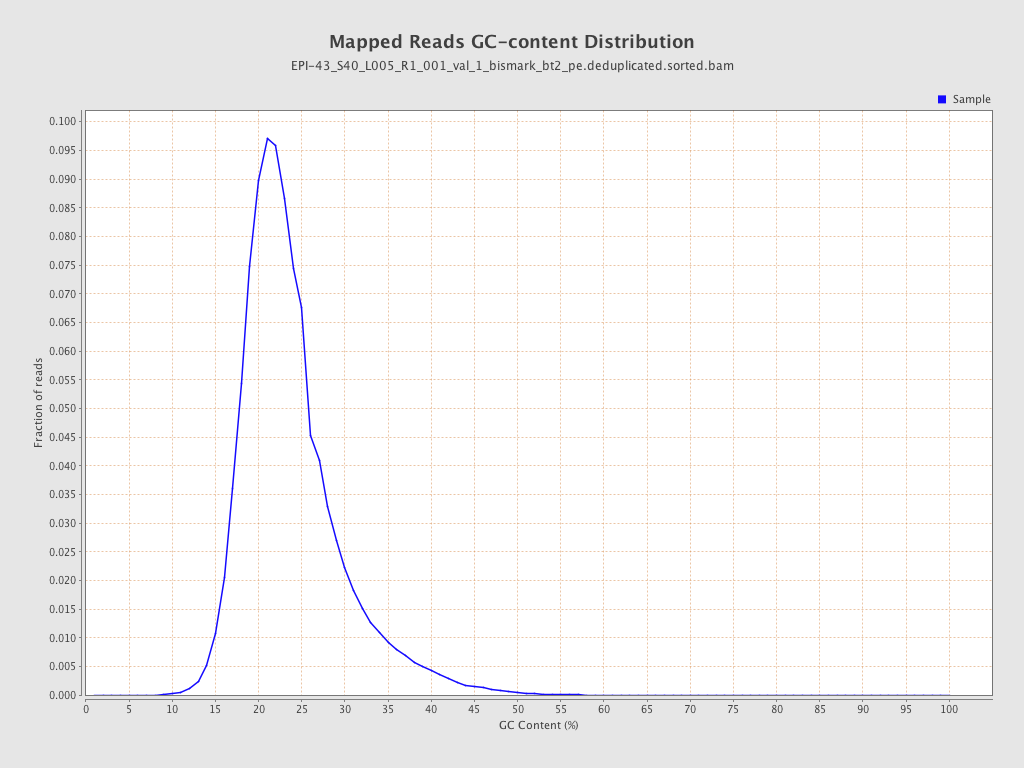
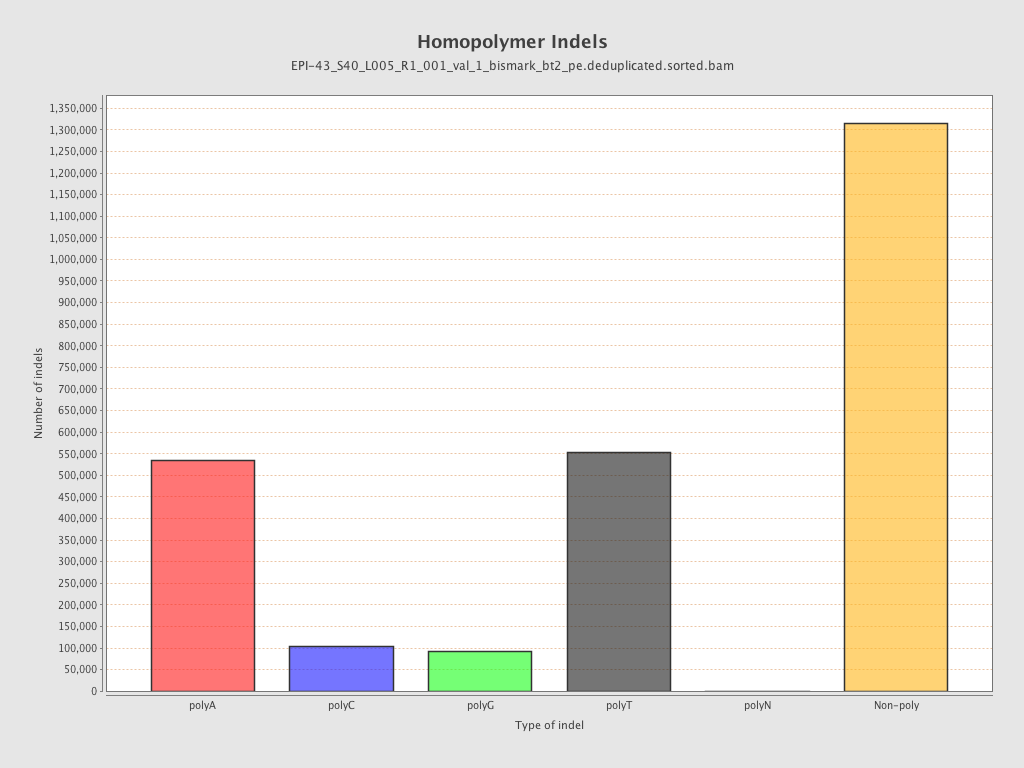
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
110923737 |
1.2374 |
3.5819 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
88078763 |
1.2656 |
4.7851 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
70214937 |
1.216 |
3.9585 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
75718880 |
1.1598 |
5.1672 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
67763682 |
1.0077 |
3.7582 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
66467357 |
1.0762 |
5.1128 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
49133874 |
1.1395 |
3.6831 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
71407683 |
1.1677 |
4.0529 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
49236385 |
1.2762 |
8.9142 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
58763839 |
1.089 |
3.4535 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
52502550 |
1.0205 |
4.2709 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
62086635 |
1.2309 |
4.9785 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
51164360 |
1.1524 |
4.0953 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
66848634 |
1.4727 |
17.7307 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
45300185 |
0.945 |
3.6689 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
38773954 |
1.2124 |
9.662 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
41141867 |
1.1781 |
4.0234 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
28191568 |
1.0164 |
5.5017 |