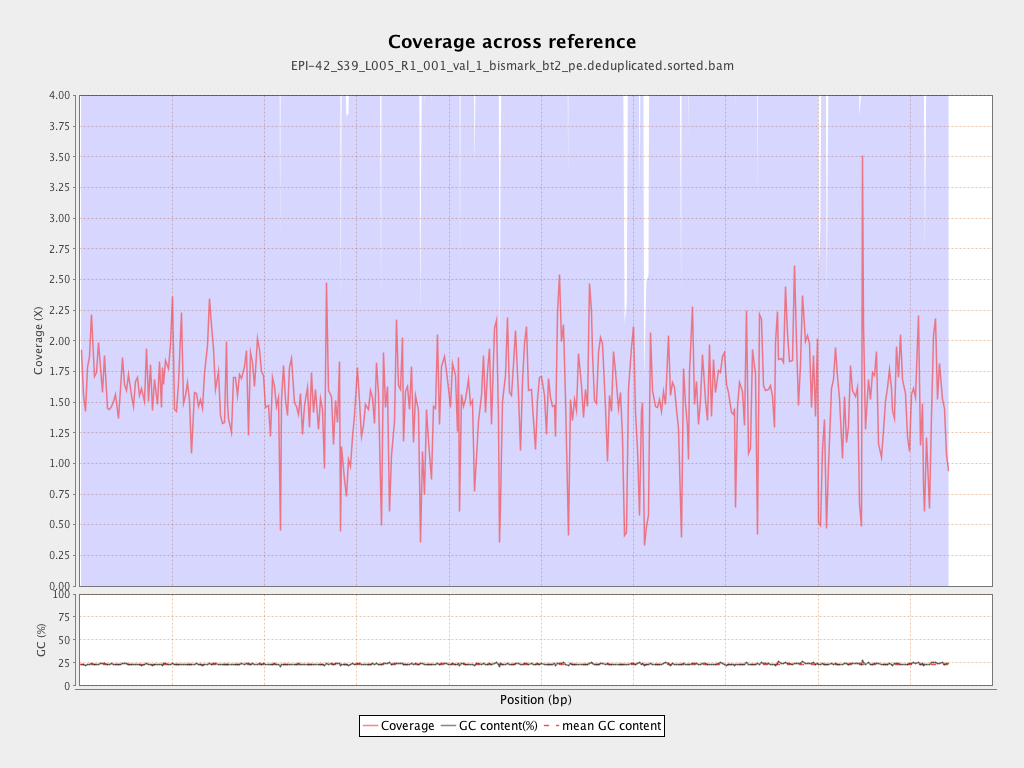
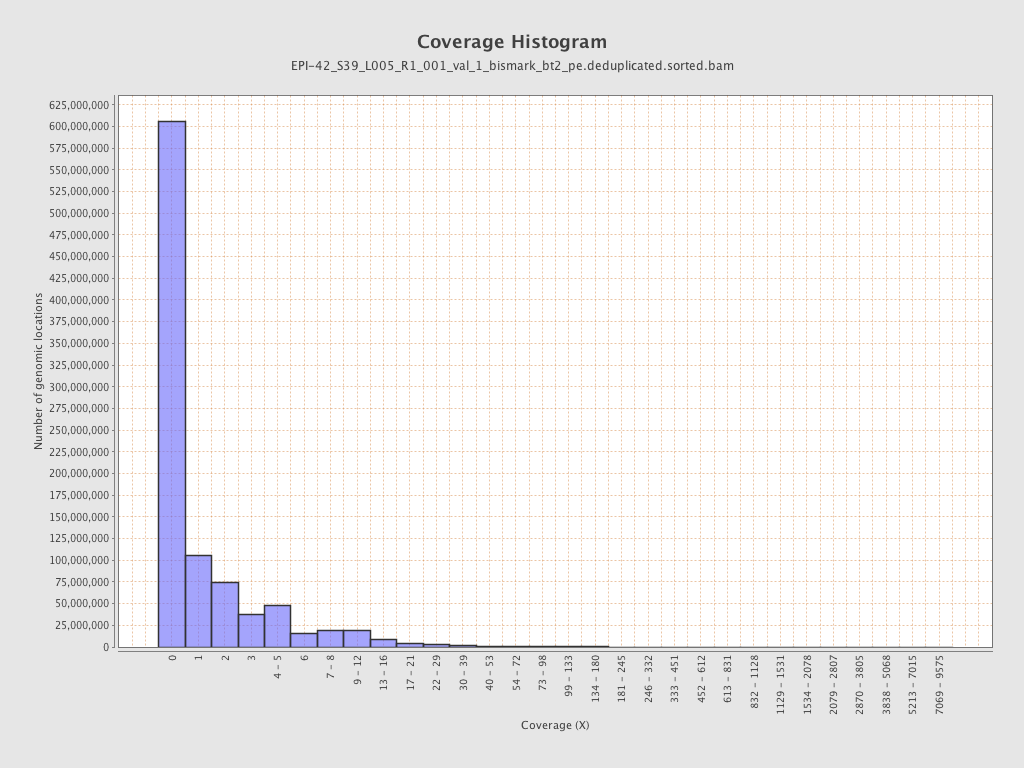
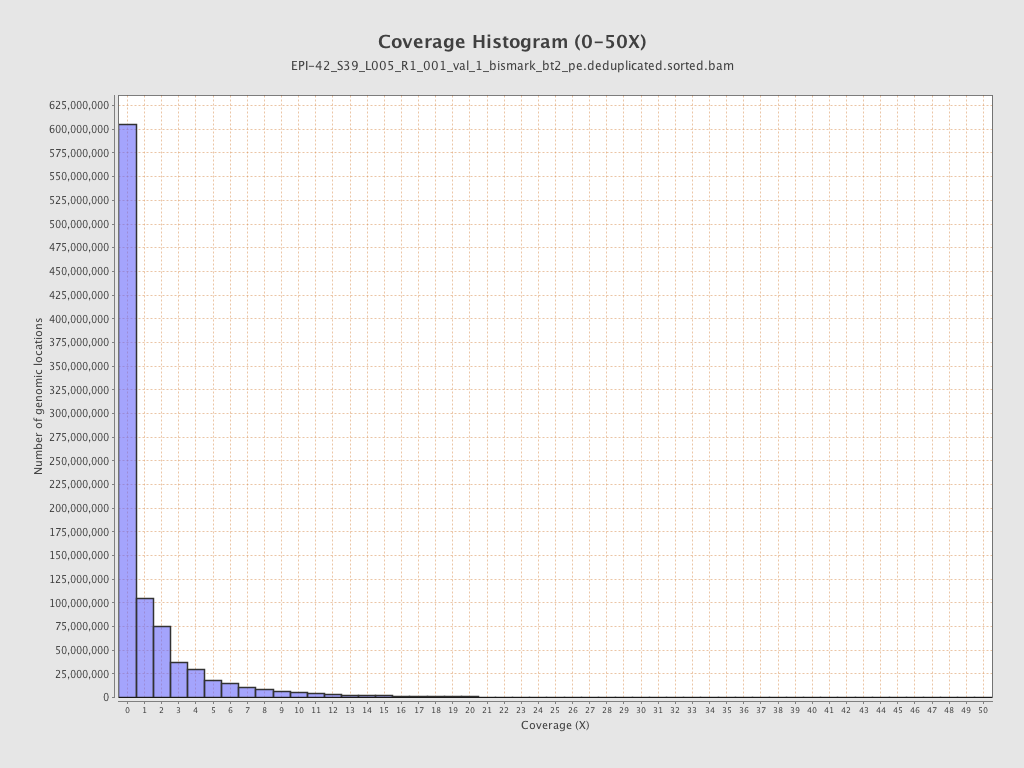
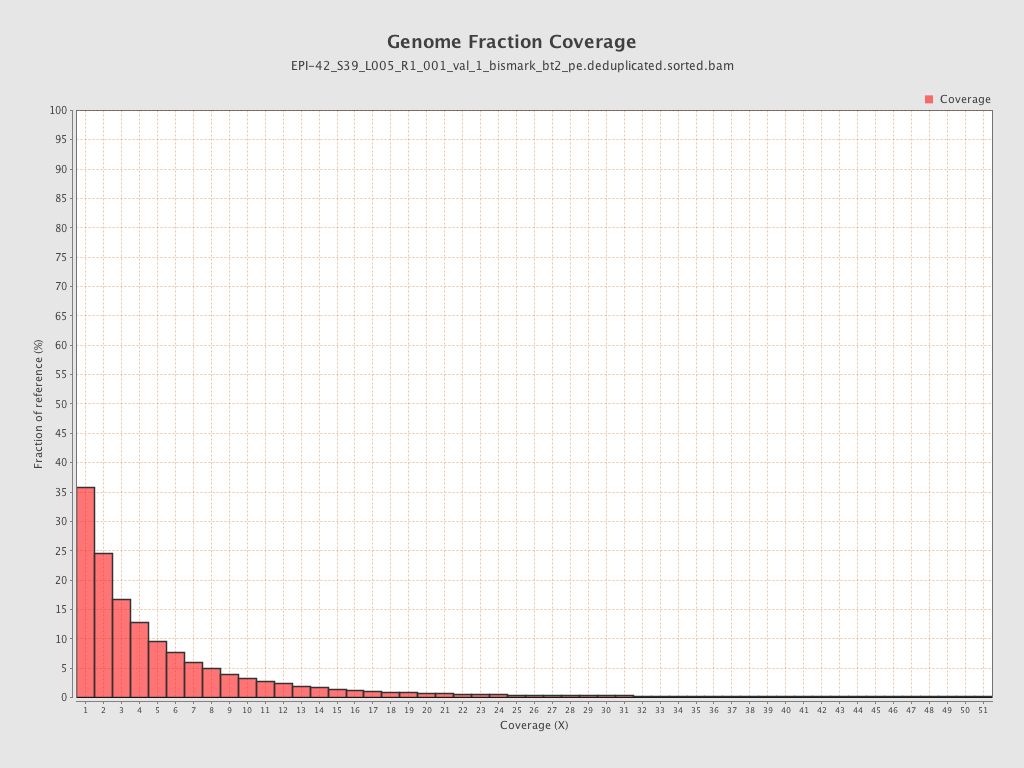
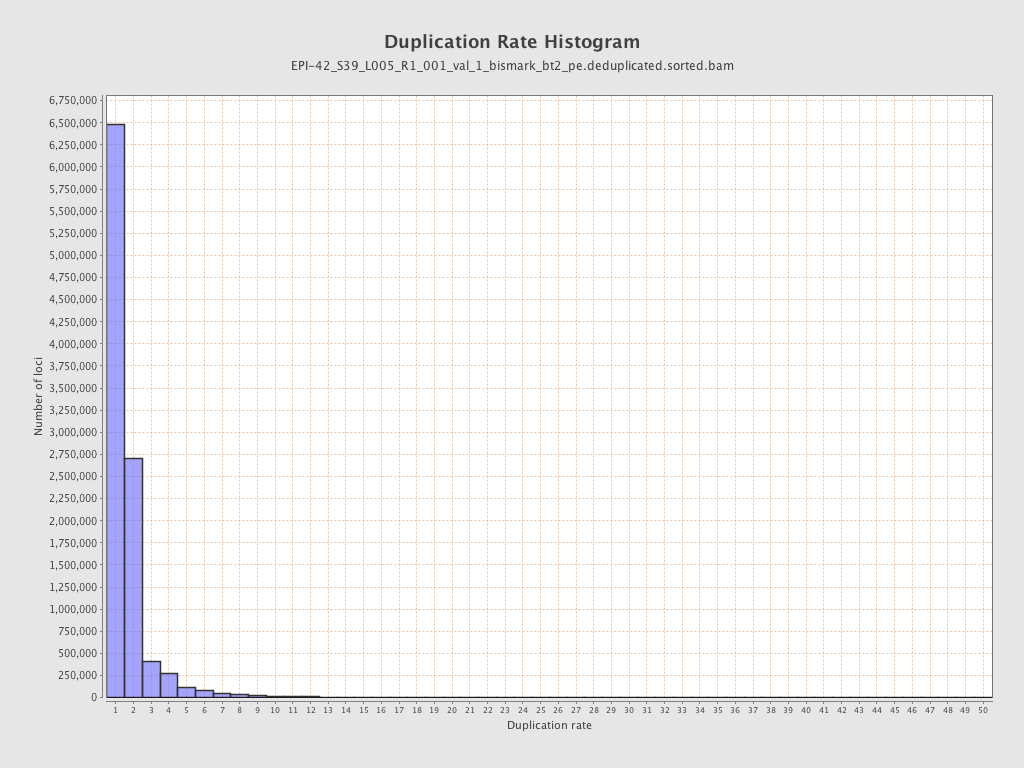
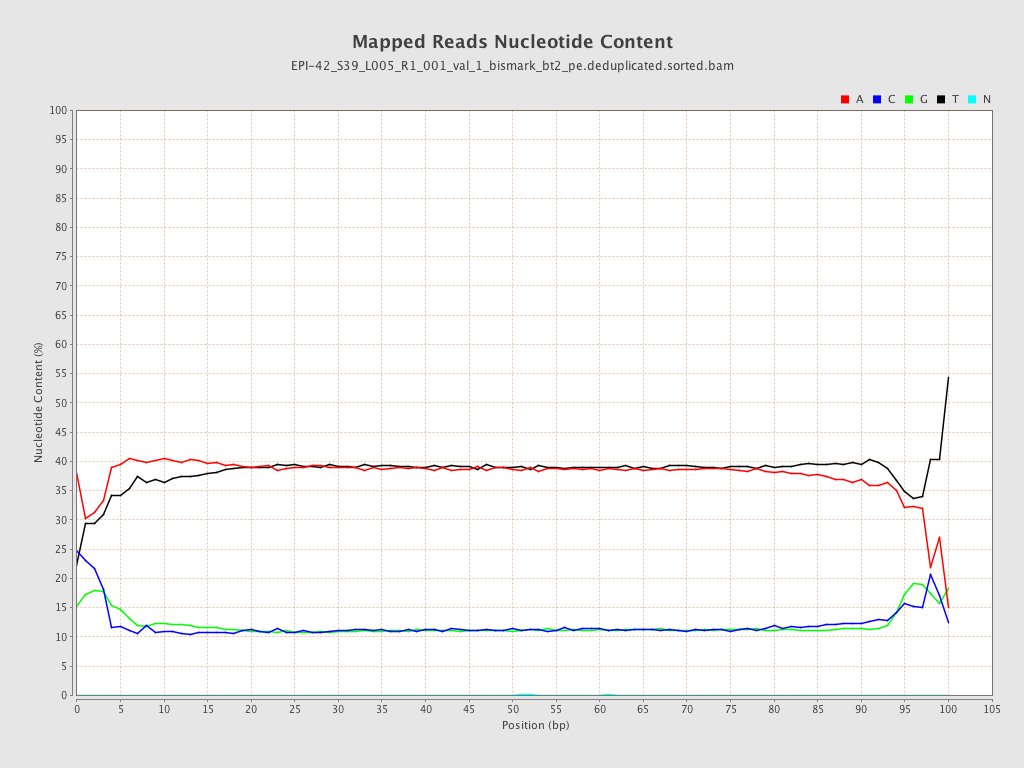
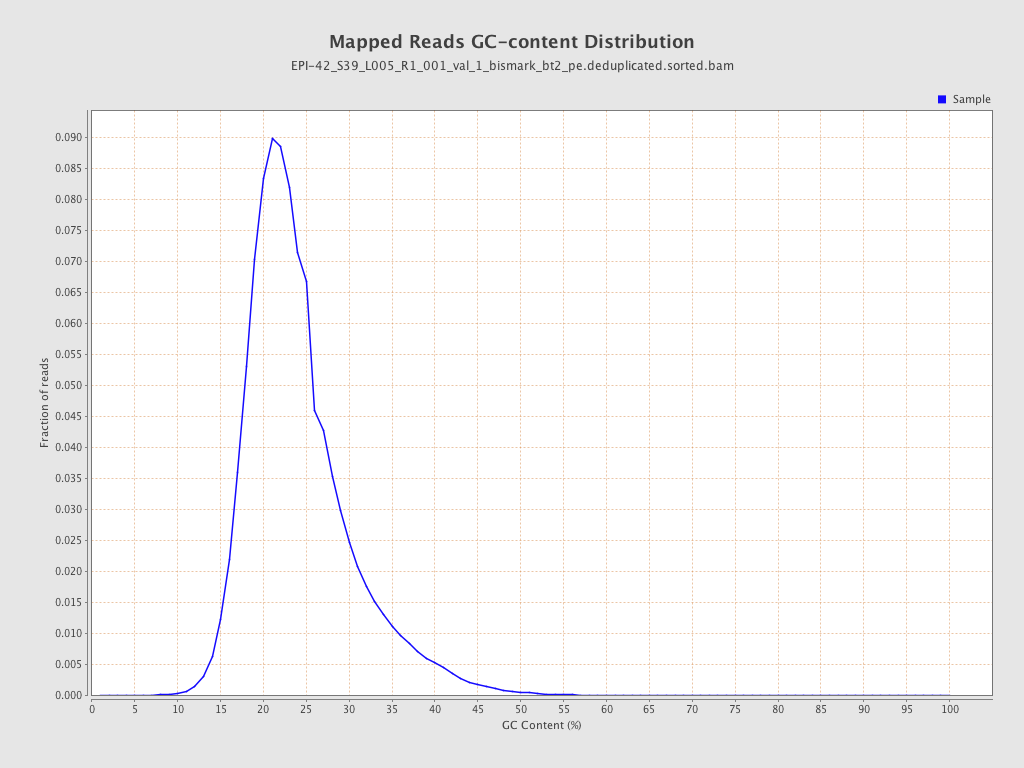
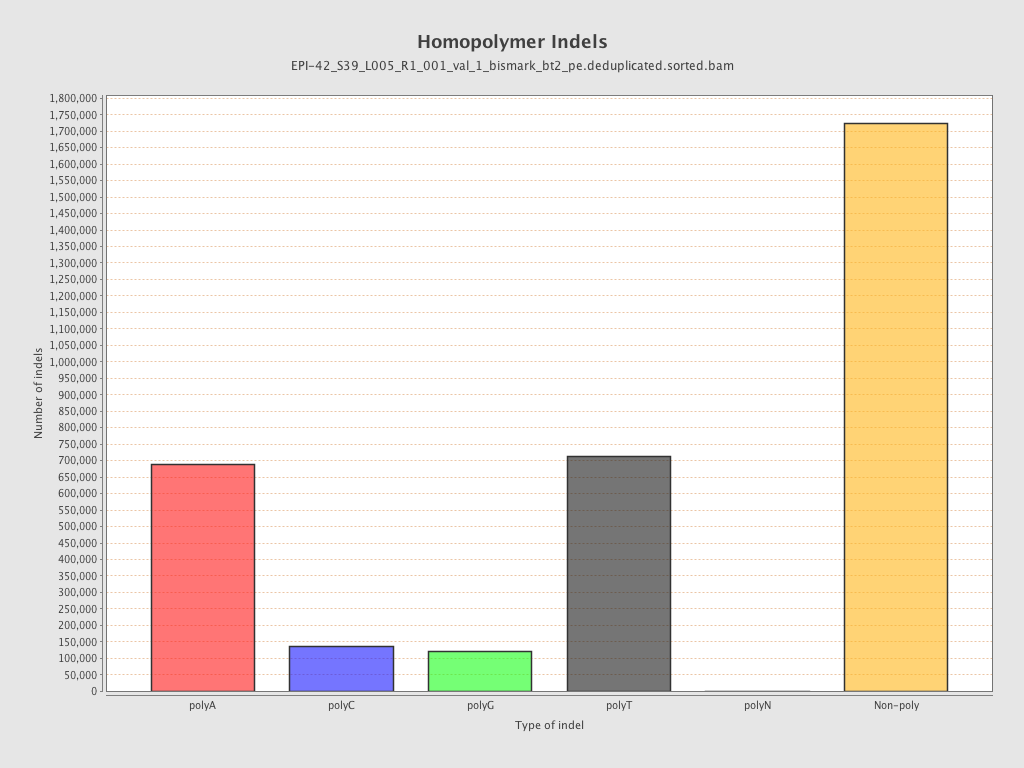
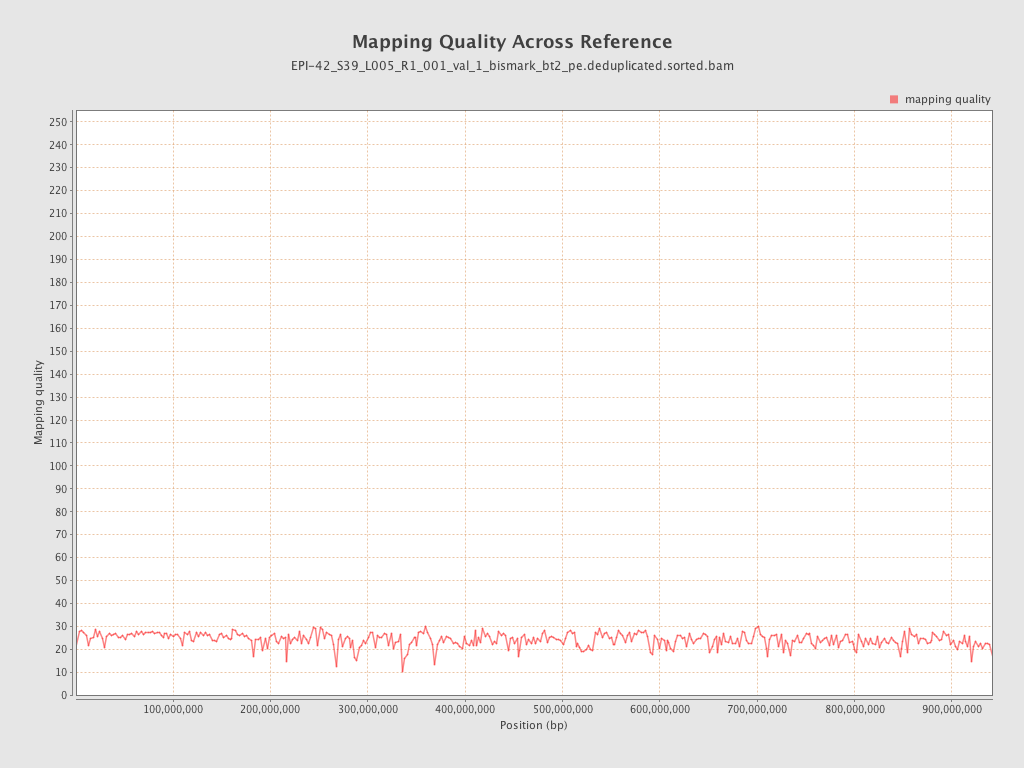
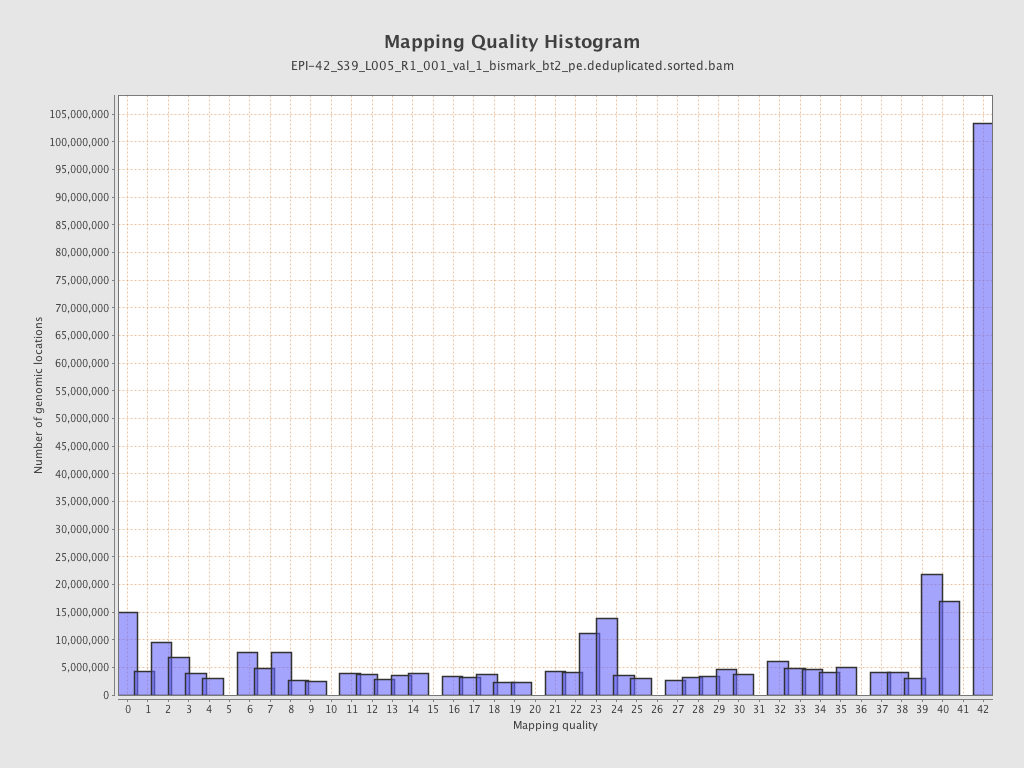
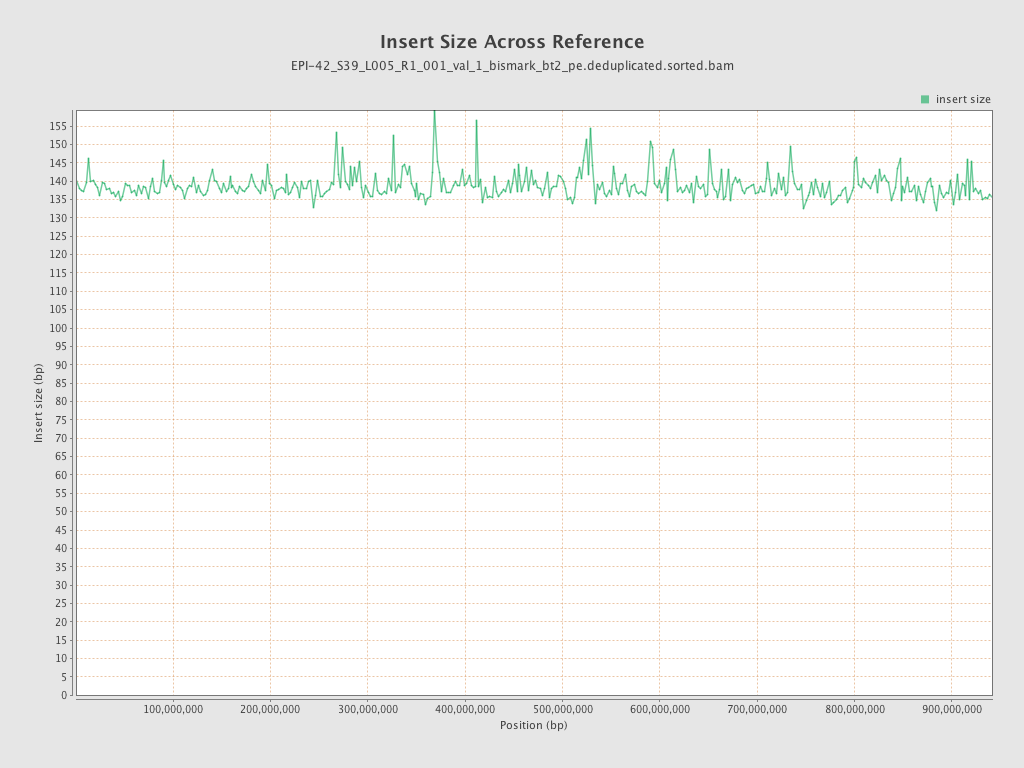
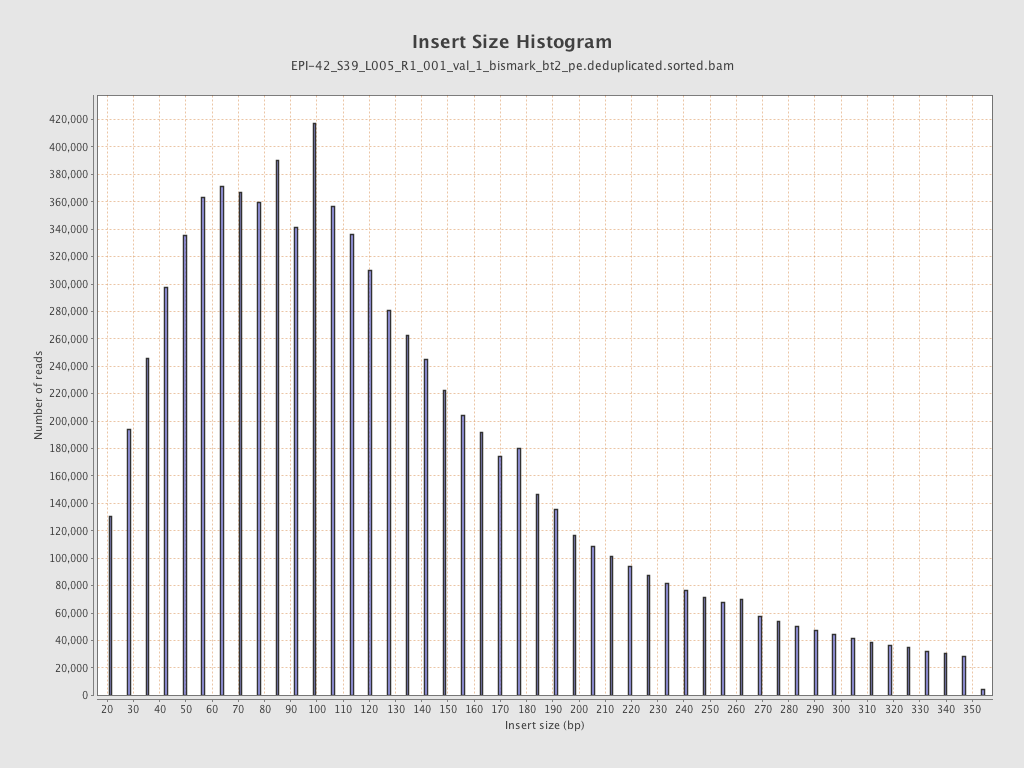
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
148334339 |
1.6547 |
4.8625 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
117317183 |
1.6857 |
5.7882 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
93866358 |
1.6256 |
5.4953 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
99768975 |
1.5281 |
6.2392 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
91025154 |
1.3536 |
5.0151 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
88749564 |
1.437 |
6.354 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
65885782 |
1.528 |
4.9073 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
96292386 |
1.5747 |
5.5483 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
65416209 |
1.6955 |
10.1087 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
78947286 |
1.463 |
4.5809 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
70125807 |
1.363 |
5.514 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
83433409 |
1.6542 |
6.3576 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
68442824 |
1.5416 |
5.7212 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
88154175 |
1.942 |
16.3117 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
60829679 |
1.2689 |
4.963 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
50944821 |
1.593 |
9.231 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
54724207 |
1.567 |
5.5021 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
38285751 |
1.3803 |
7.1496 |