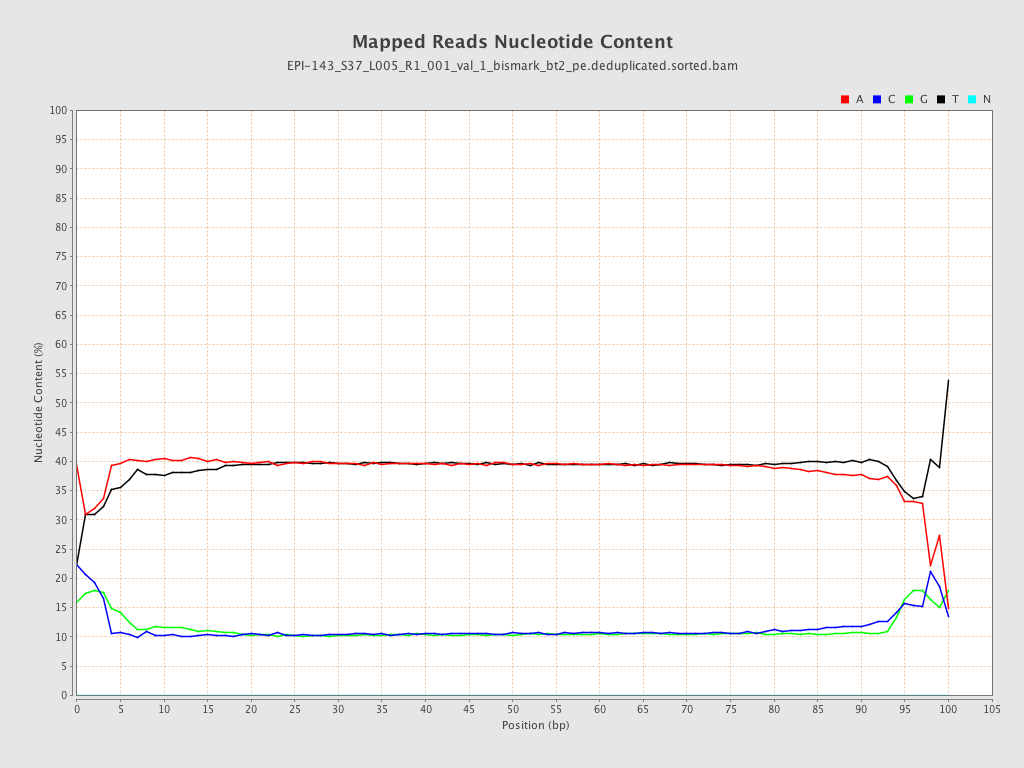
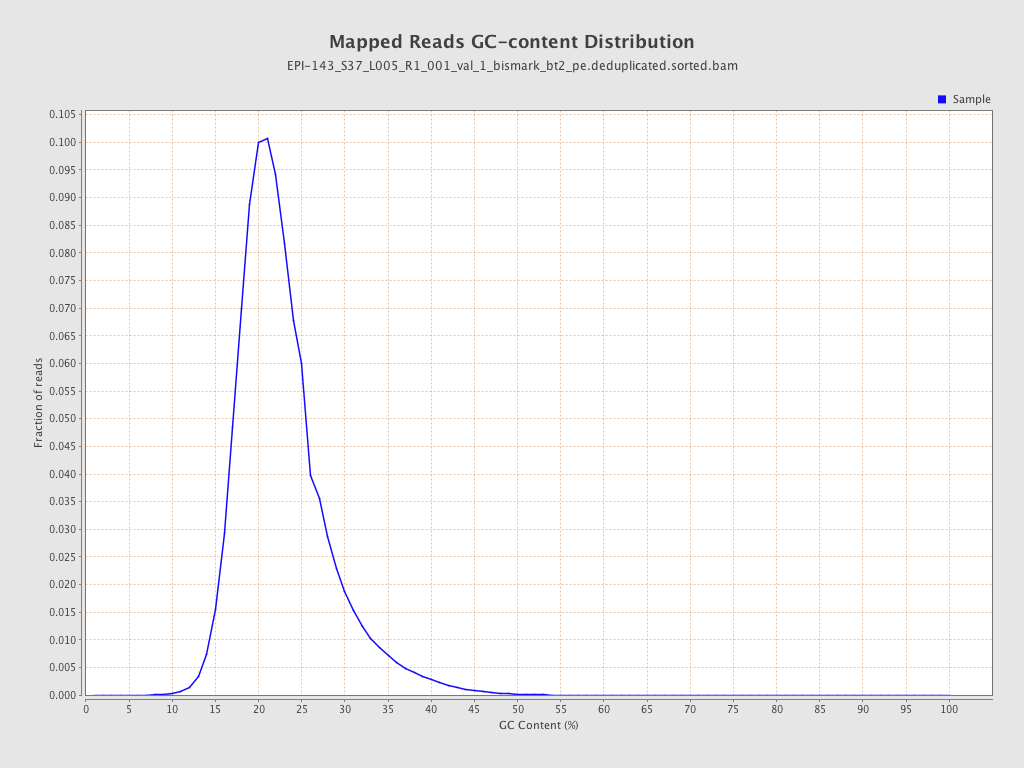
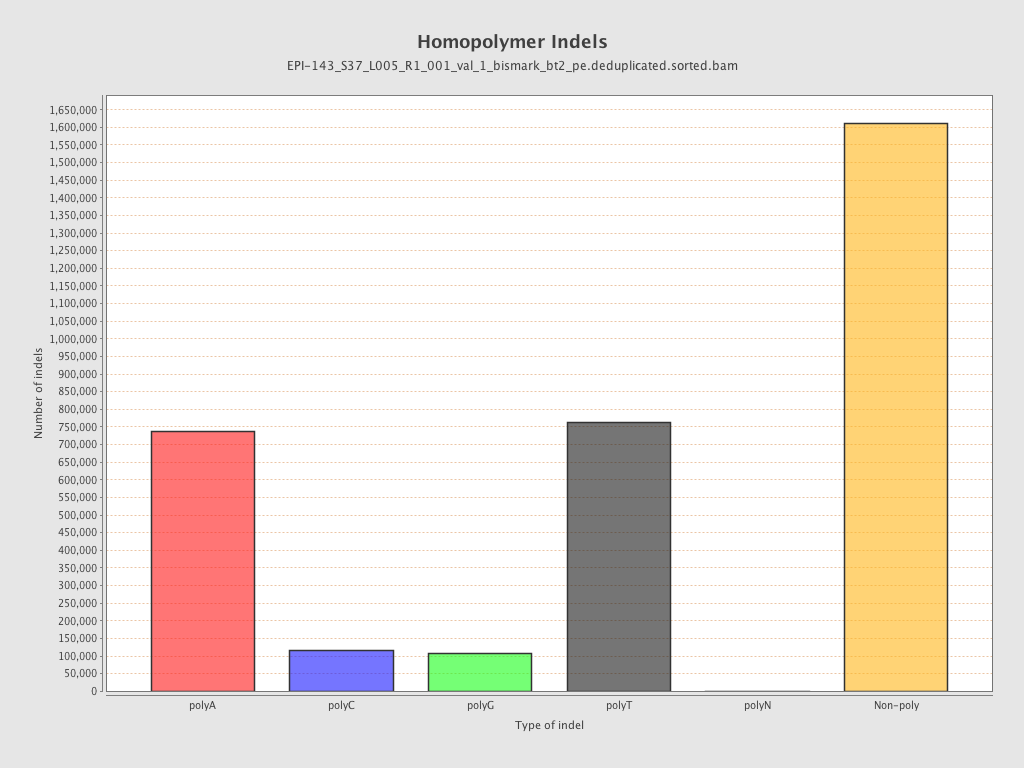
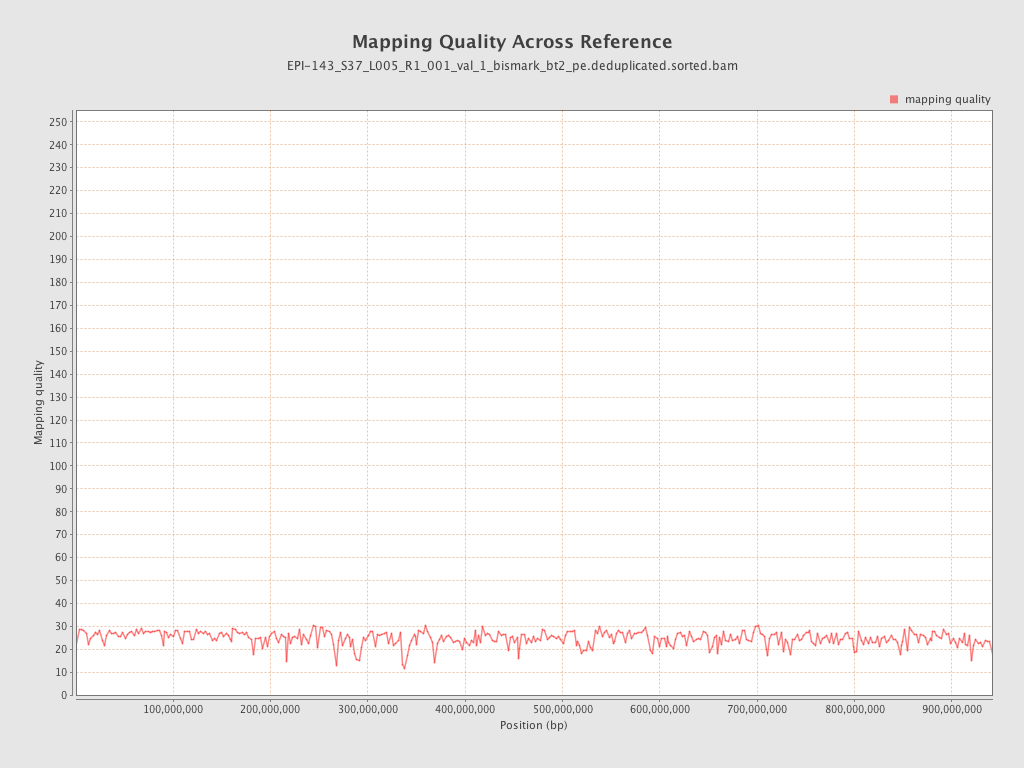
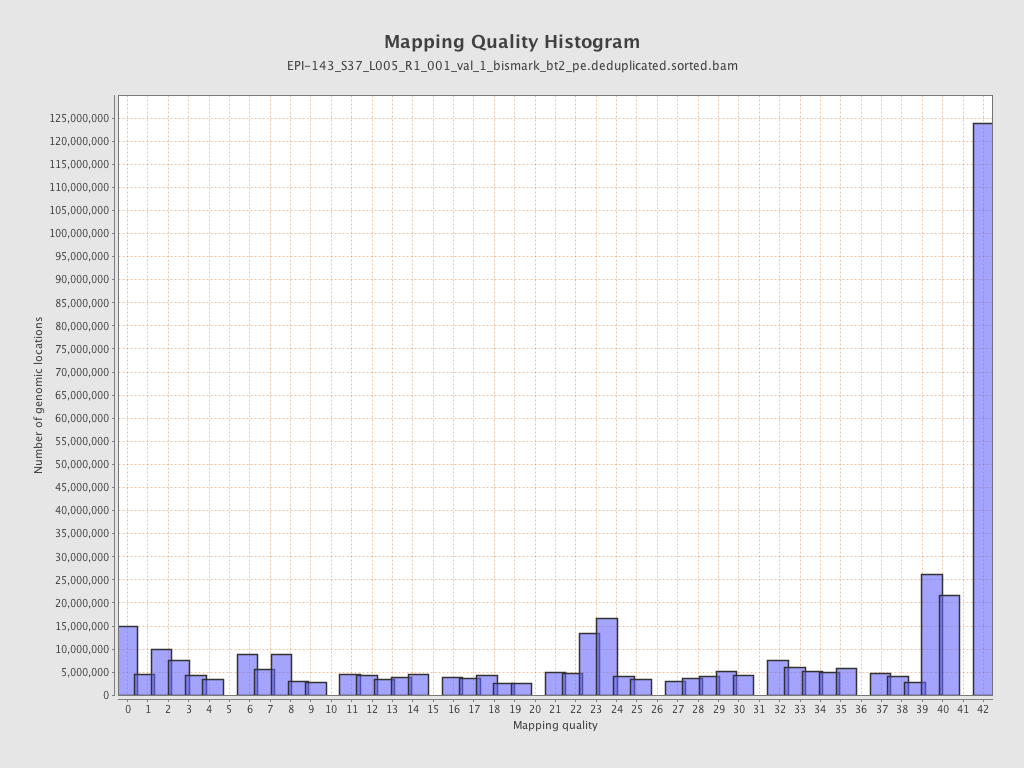
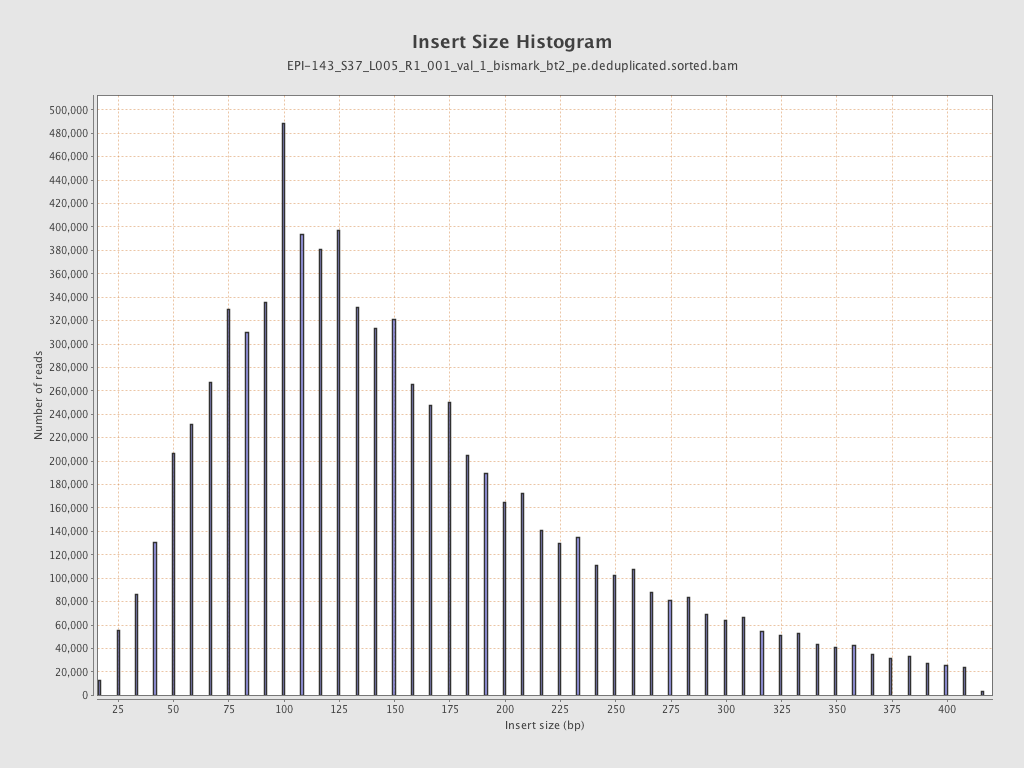
| Name |
Length |
Mapped bases |
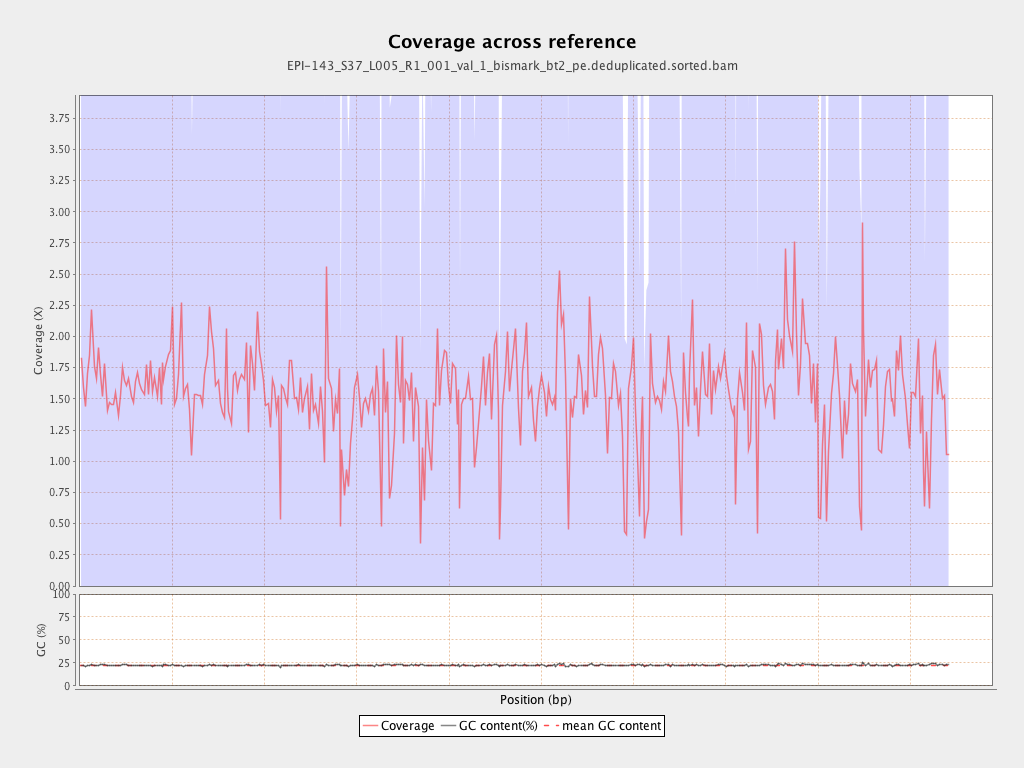
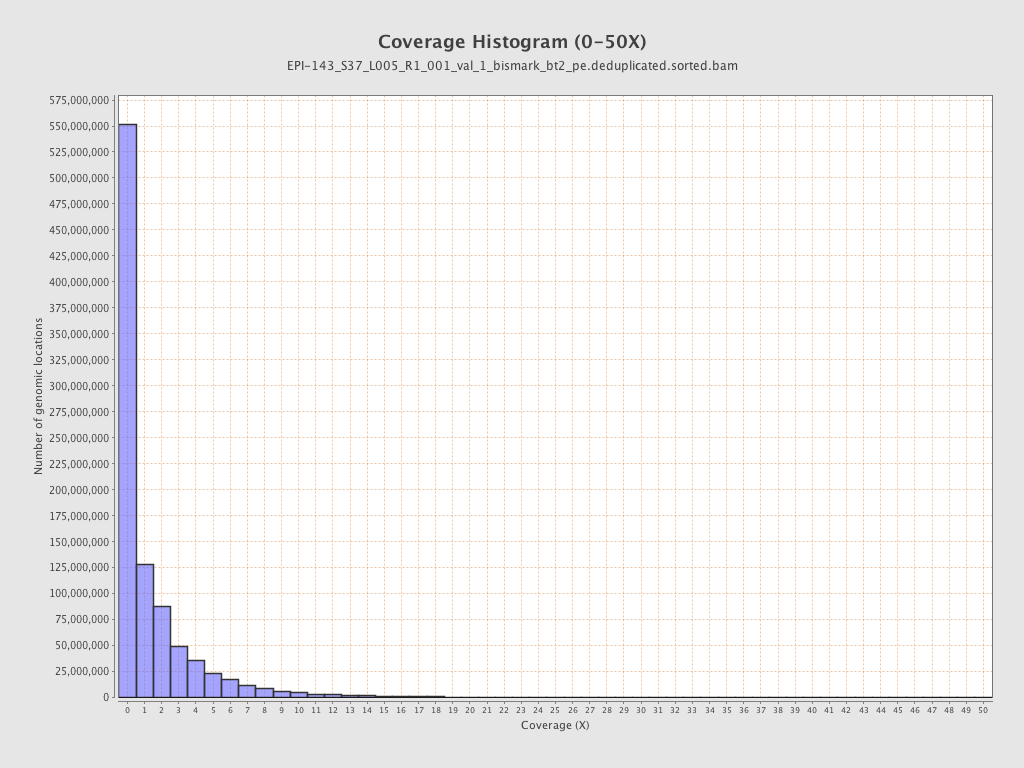
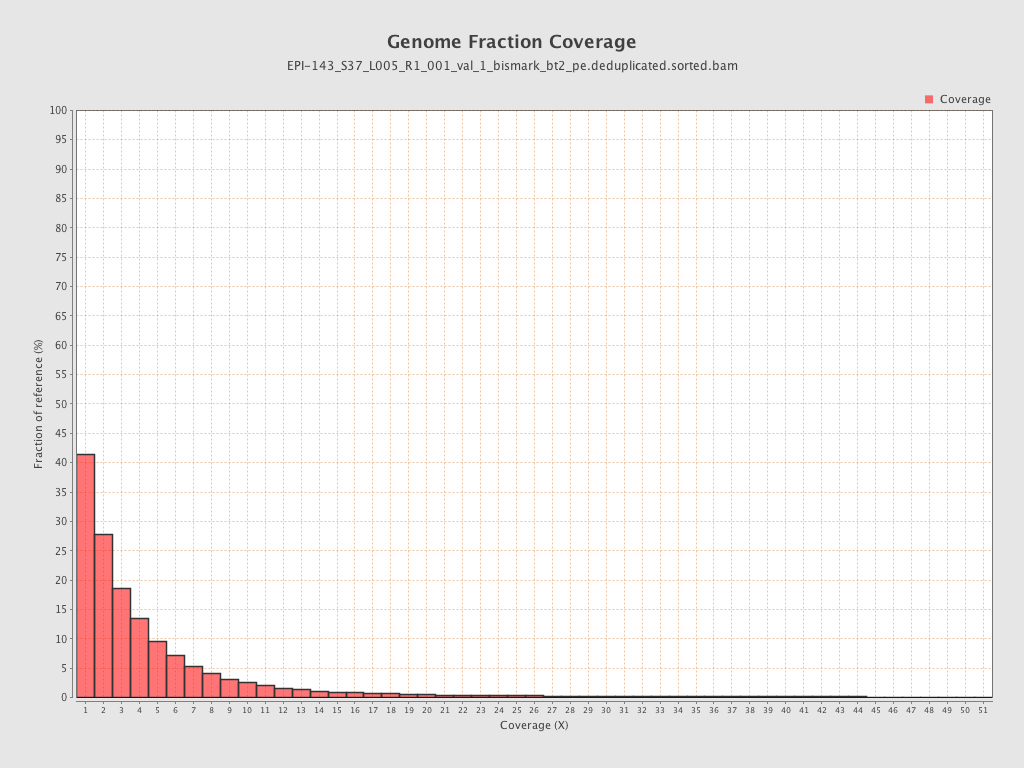
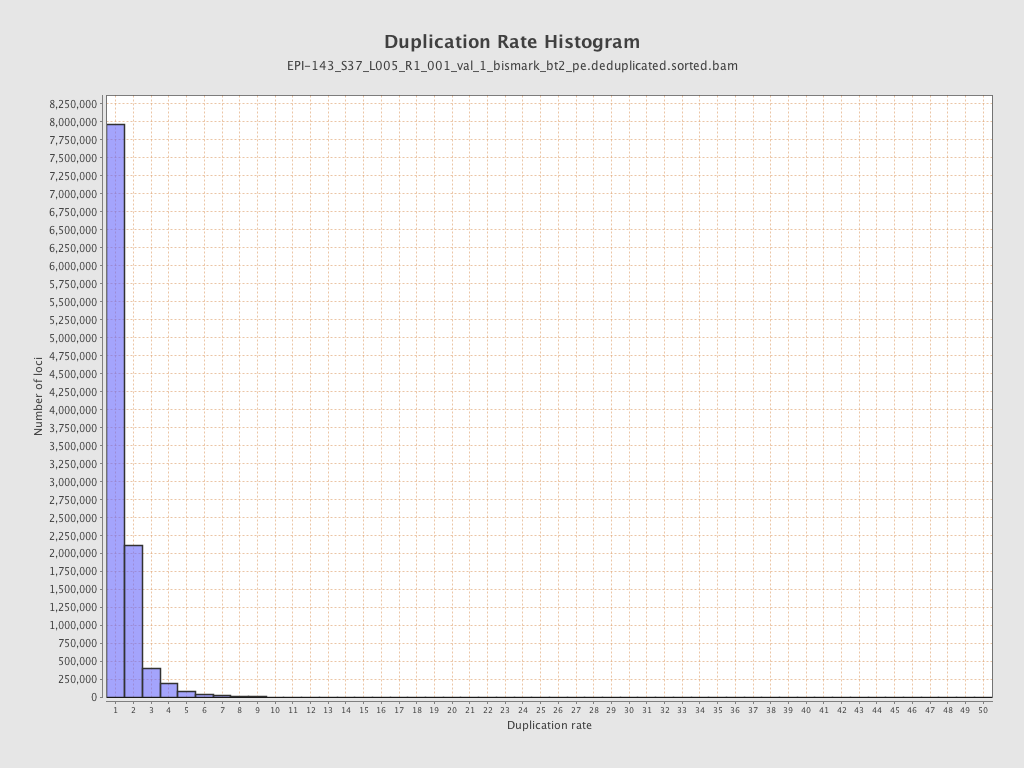
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
146360651 |
1.6327 |
4.0863 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
116447567 |
1.6732 |
5.1603 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
93493204 |
1.6191 |
4.7259 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
100457098 |
1.5387 |
6.1988 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
90254303 |
1.3421 |
4.5261 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
89050779 |
1.4419 |
5.6967 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
65326708 |
1.515 |
4.4024 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
95206149 |
1.5569 |
5.0773 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
65724963 |
1.7035 |
11.1391 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
77319821 |
1.4329 |
3.7948 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
71304075 |
1.3859 |
5.0169 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
82626649 |
1.6382 |
5.9168 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
67545773 |
1.5214 |
5.0823 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
87981398 |
1.9382 |
19.4421 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
61894376 |
1.2911 |
4.3099 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
51072527 |
1.597 |
8.0597 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
54256813 |
1.5536 |
4.4129 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
37427589 |
1.3494 |
5.4659 |