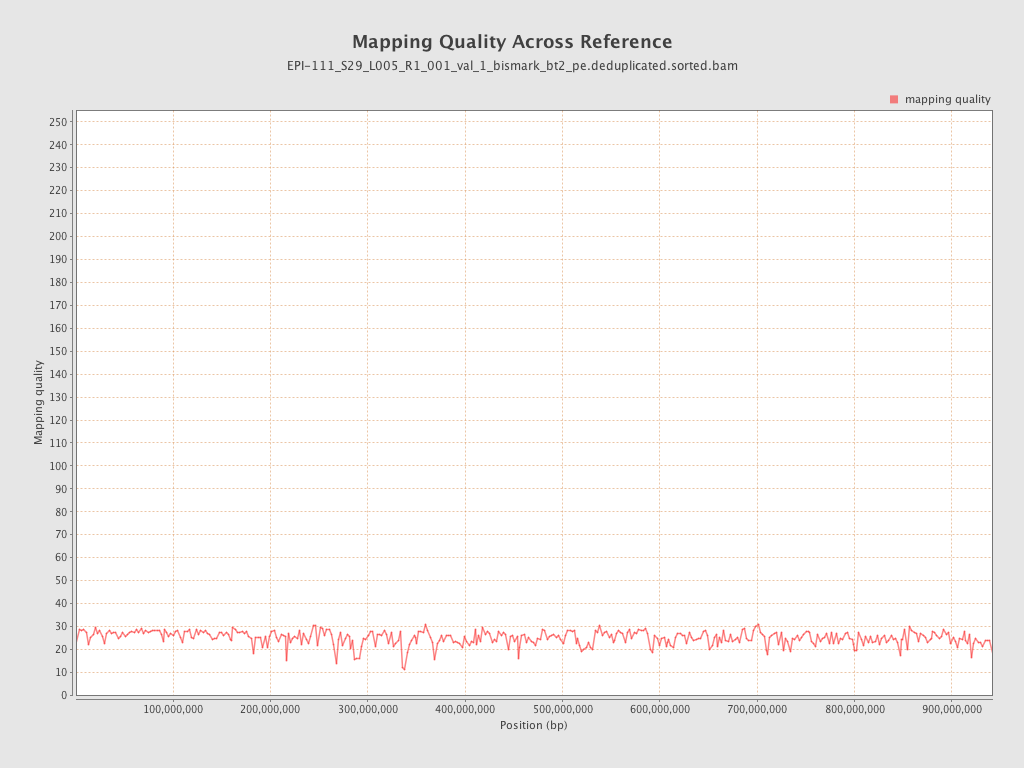
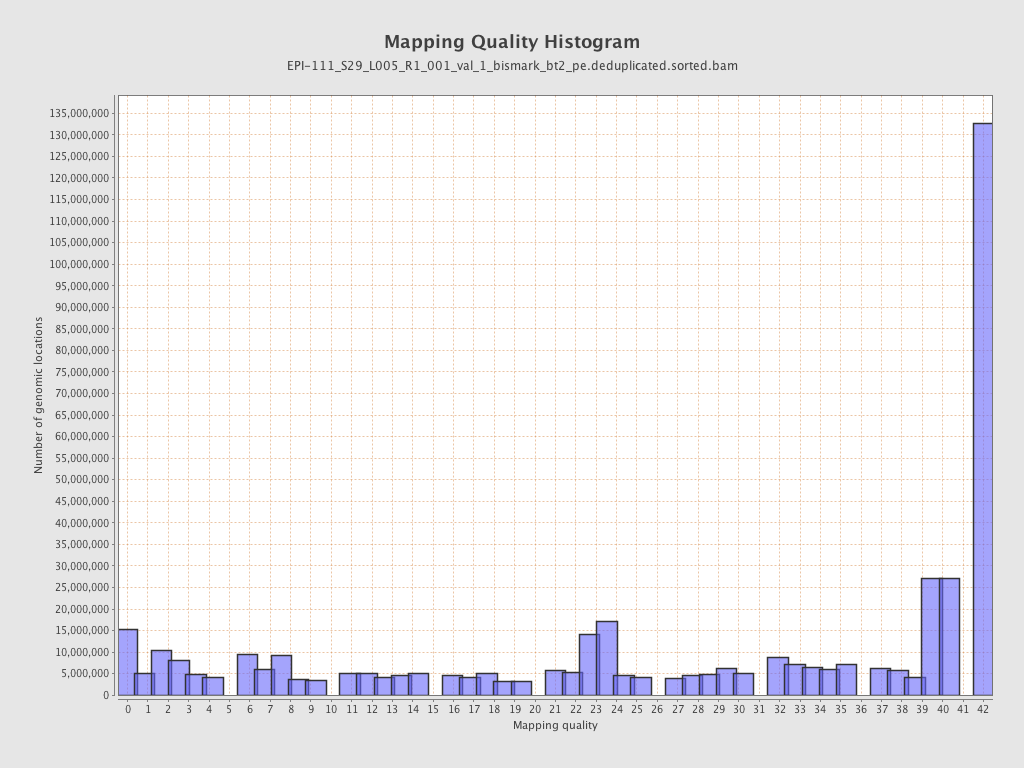
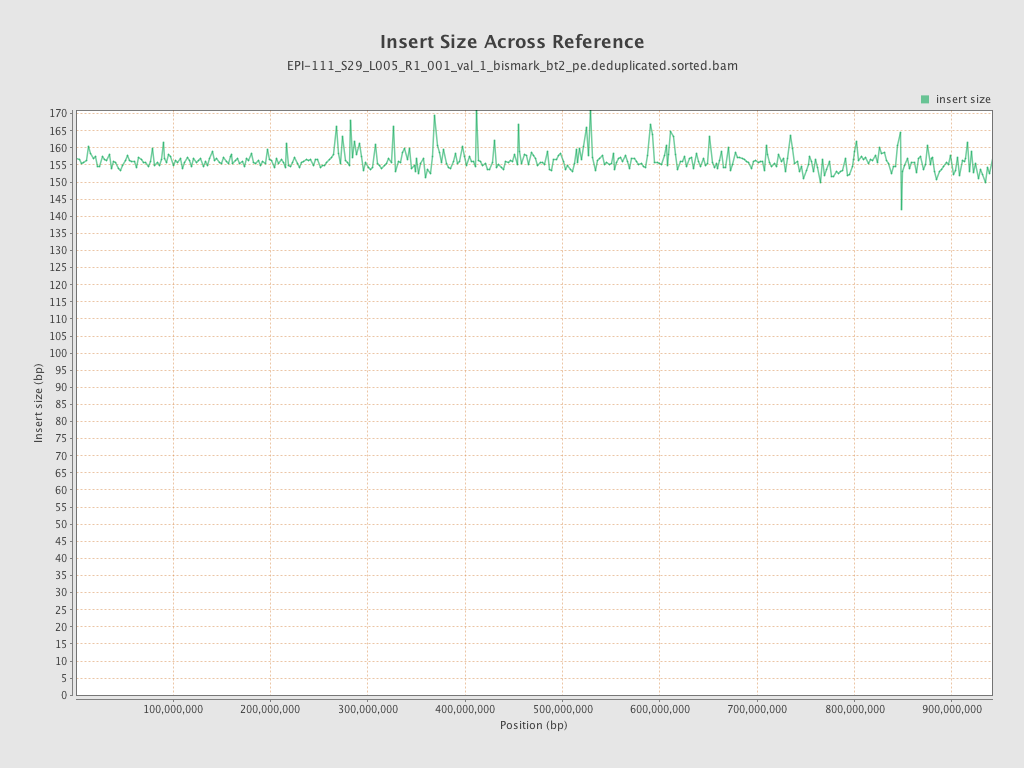
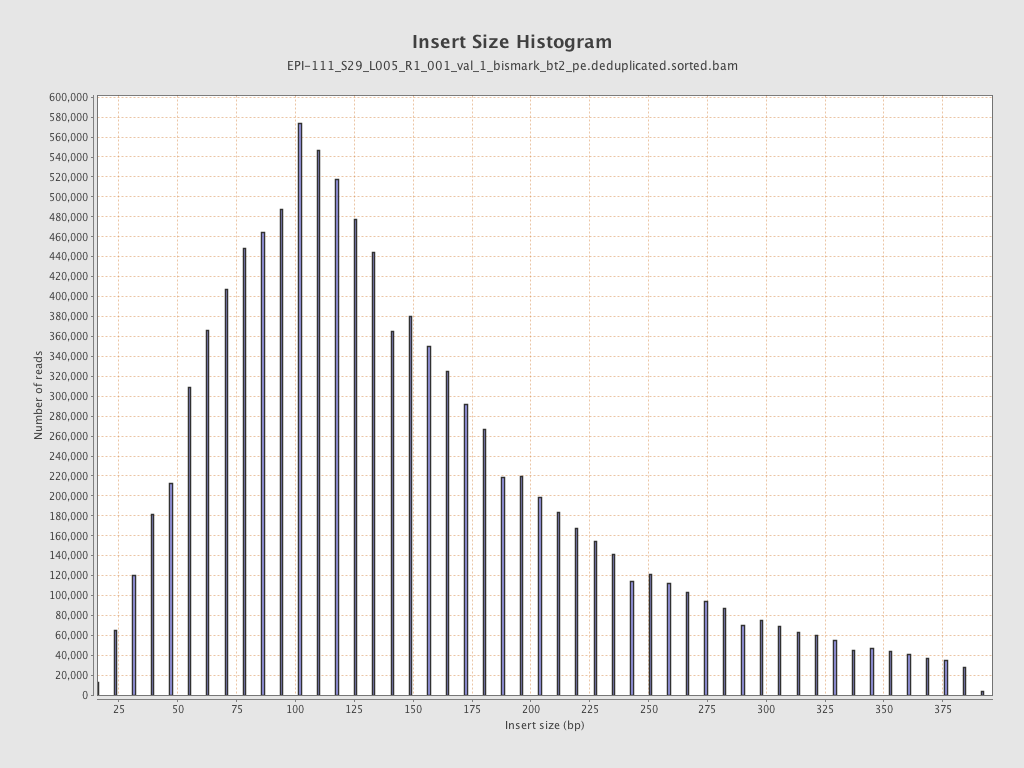
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
194679434 |
2.1717 |
5.1333 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
153602044 |
2.207 |
6.3942 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
121866655 |
2.1105 |
5.8421 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
133747673 |
2.0486 |
7.3313 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
119227350 |
1.7729 |
5.3531 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
117492491 |
1.9024 |
7.0934 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
85752964 |
1.9887 |
5.467 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
124391221 |
2.0342 |
5.9635 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
84226478 |
2.1831 |
12.23 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
101985805 |
1.89 |
4.6902 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
91719013 |
1.7827 |
5.7924 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
109028220 |
2.1616 |
7.2089 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
88582152 |
1.9952 |
6.1265 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
115124433 |
2.5362 |
21.5974 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
81056376 |
1.6908 |
5.3011 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
66685102 |
2.0852 |
9.2017 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
71728706 |
2.0539 |
5.4157 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
49028149 |
1.7676 |
6.422 |