| Name |
Length |
Mapped bases |
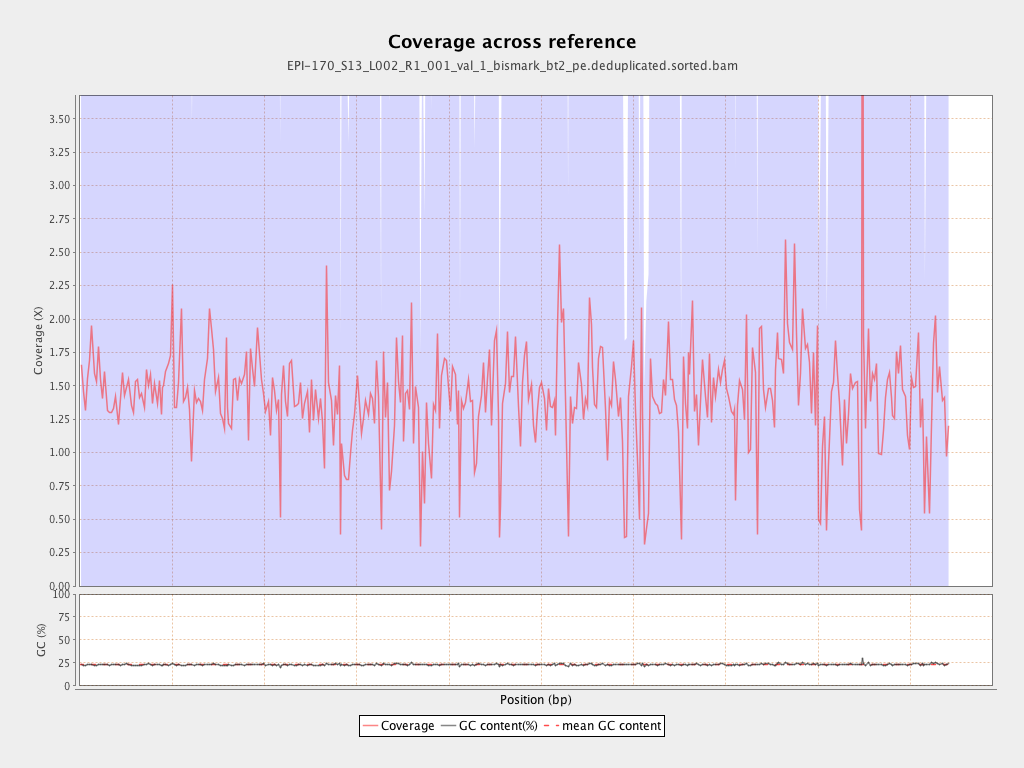
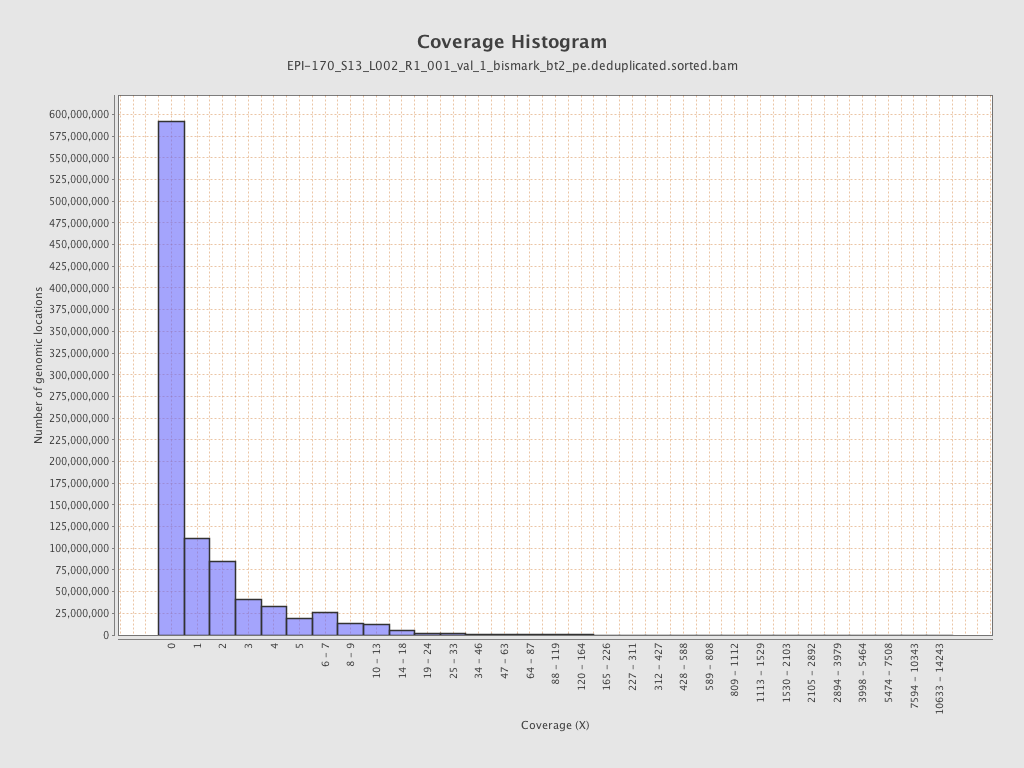
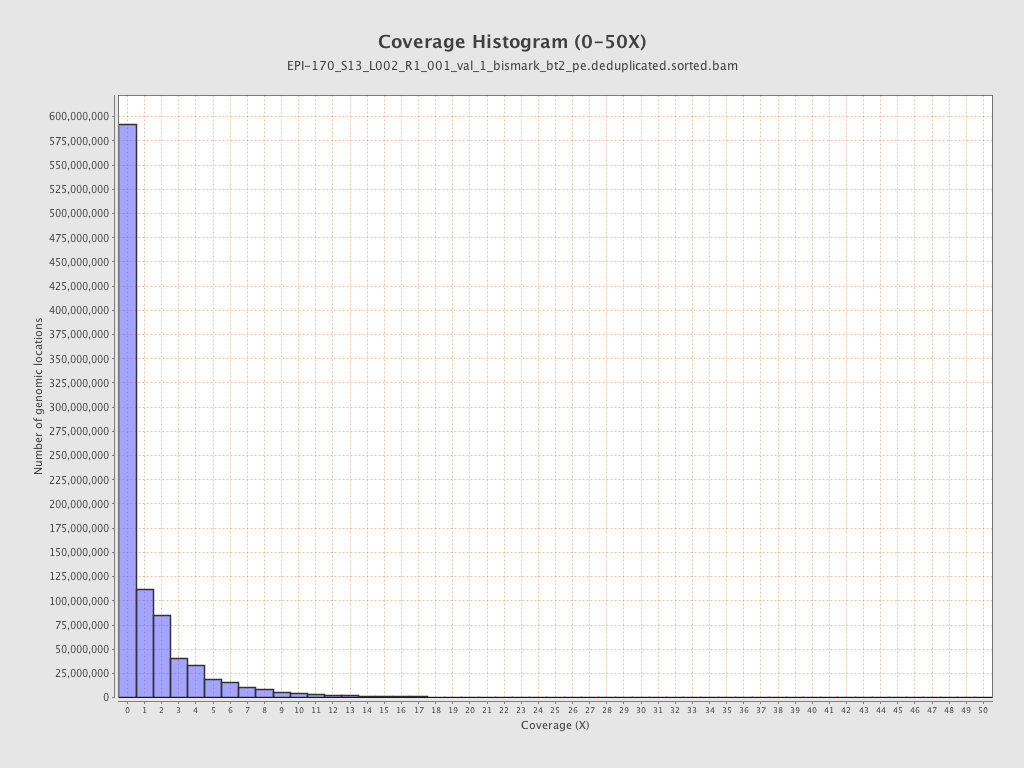
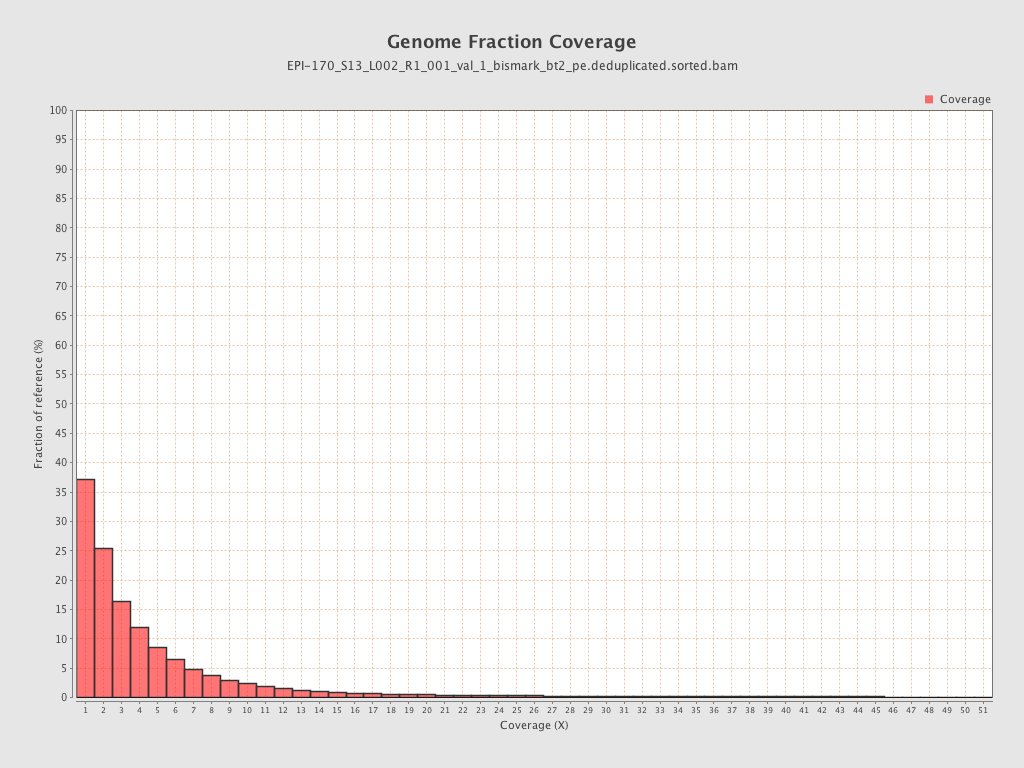
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
132387920 |
1.4768 |
5.4774 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
106711114 |
1.5333 |
7.076 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
85101351 |
1.4738 |
4.8238 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
92389185 |
1.4151 |
6.8481 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
83349158 |
1.2394 |
5.1165 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
82216032 |
1.3312 |
9.7748 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
59984706 |
1.3911 |
4.5096 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
86593833 |
1.4161 |
5.19 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
60260205 |
1.5619 |
12.7597 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
70490682 |
1.3063 |
4.1133 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
64535068 |
1.2543 |
5.5224 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
74793739 |
1.4829 |
6.5972 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
62174665 |
1.4004 |
5.6752 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
81422750 |
1.7937 |
22.4889 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
56146232 |
1.1712 |
4.6591 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
48685520 |
1.5223 |
20.2662 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
50569862 |
1.448 |
6.0563 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
35774190 |
1.2897 |
8.0387 |