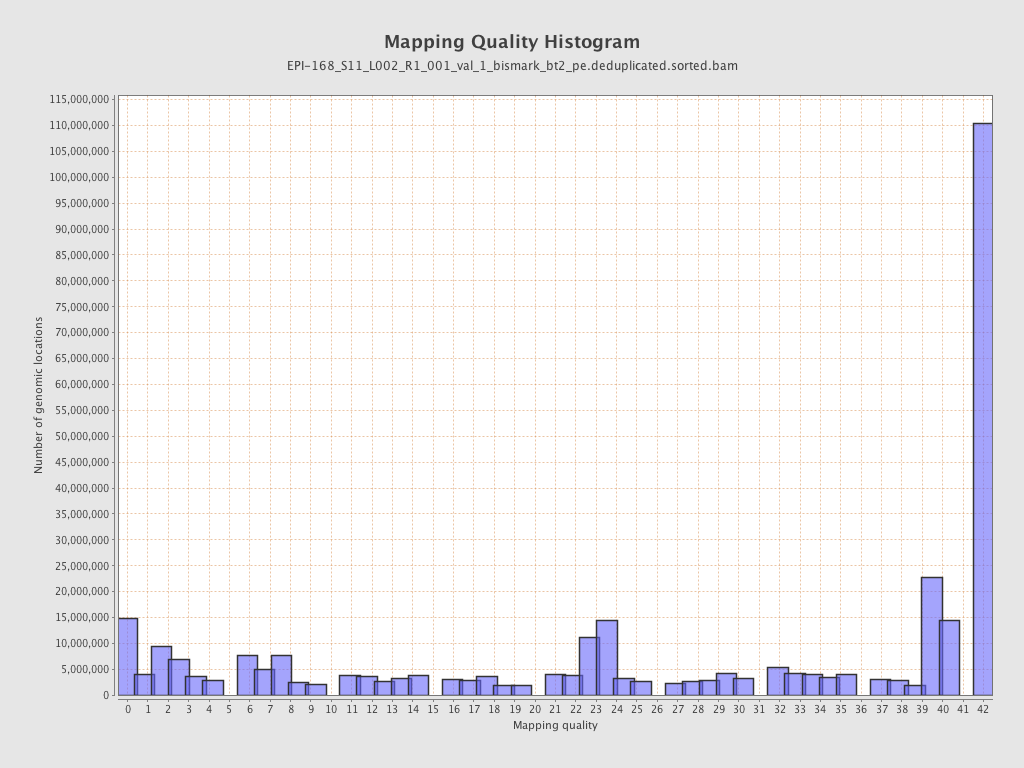
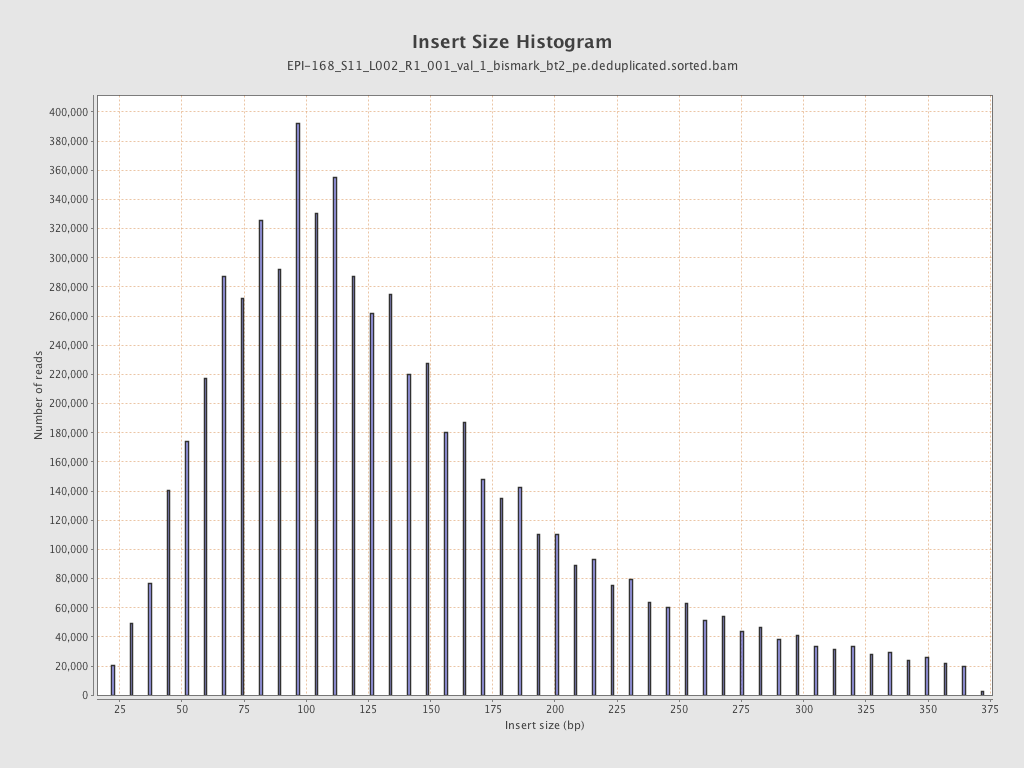
| Name |
Length |
Mapped bases |
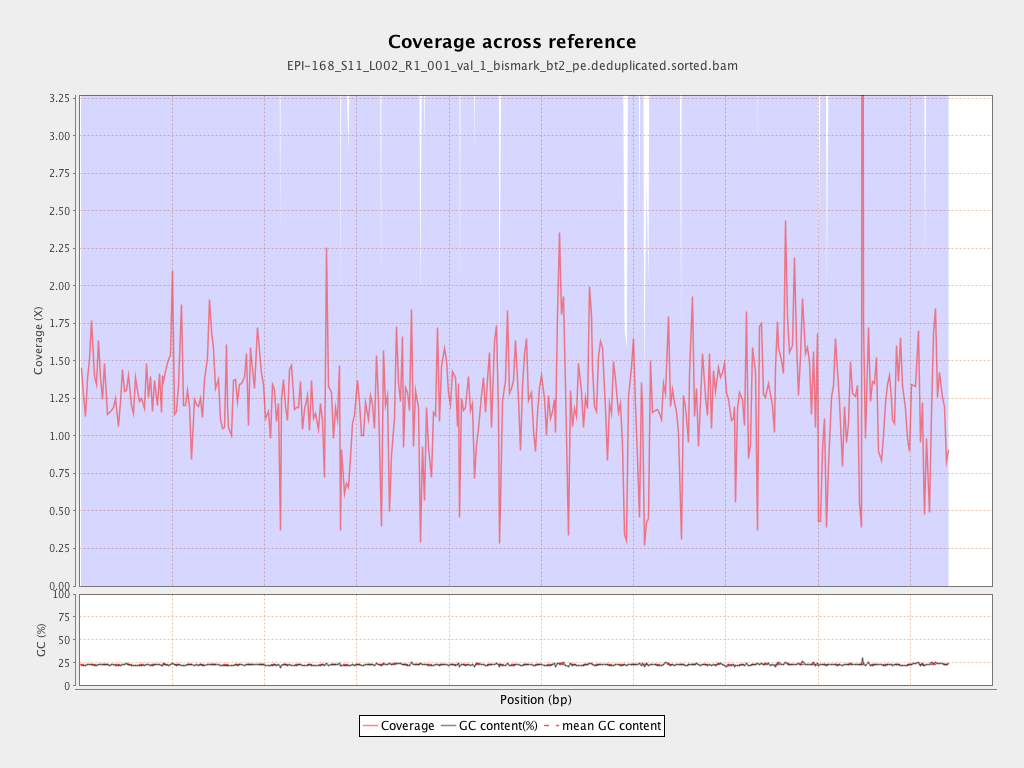
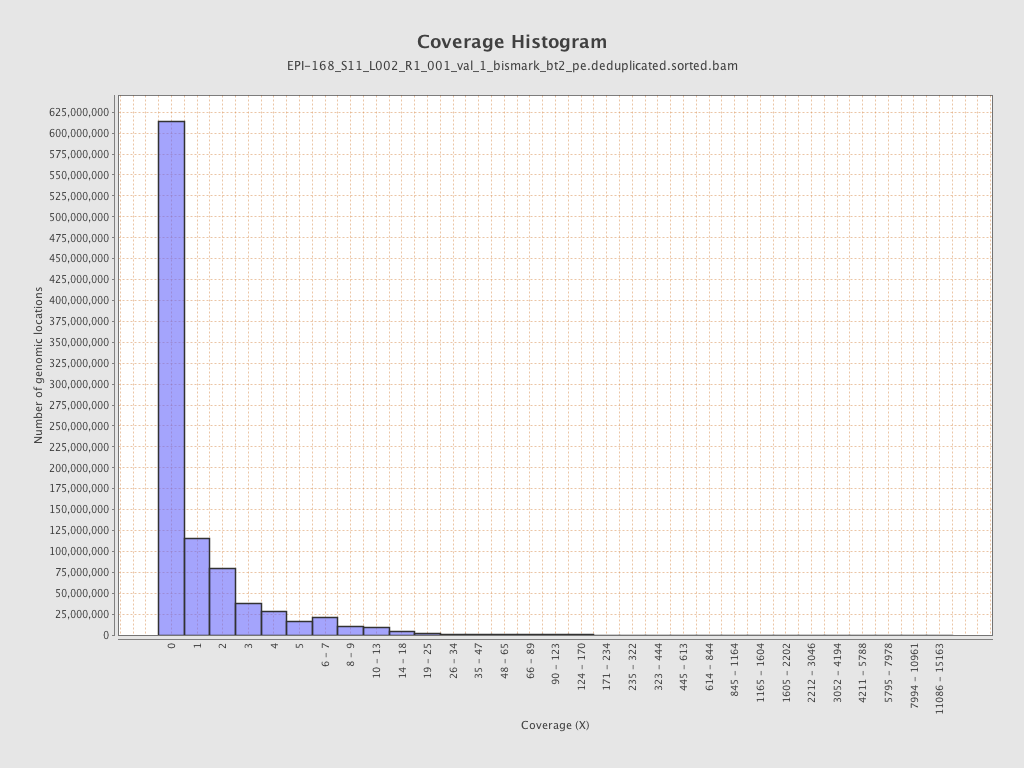
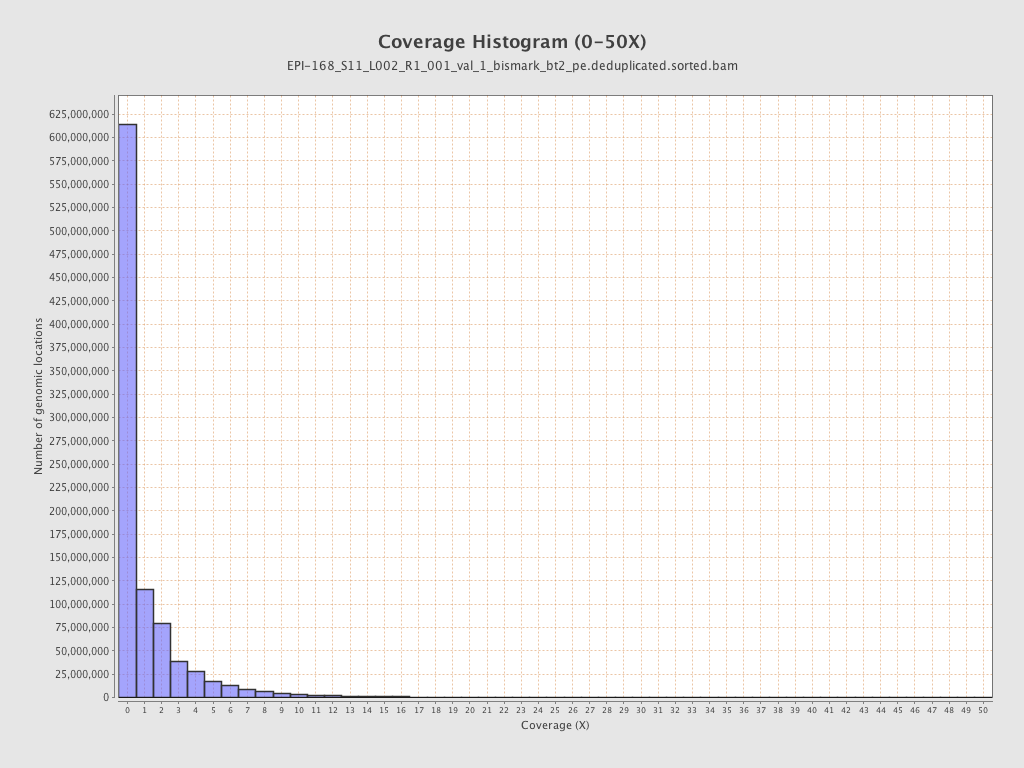
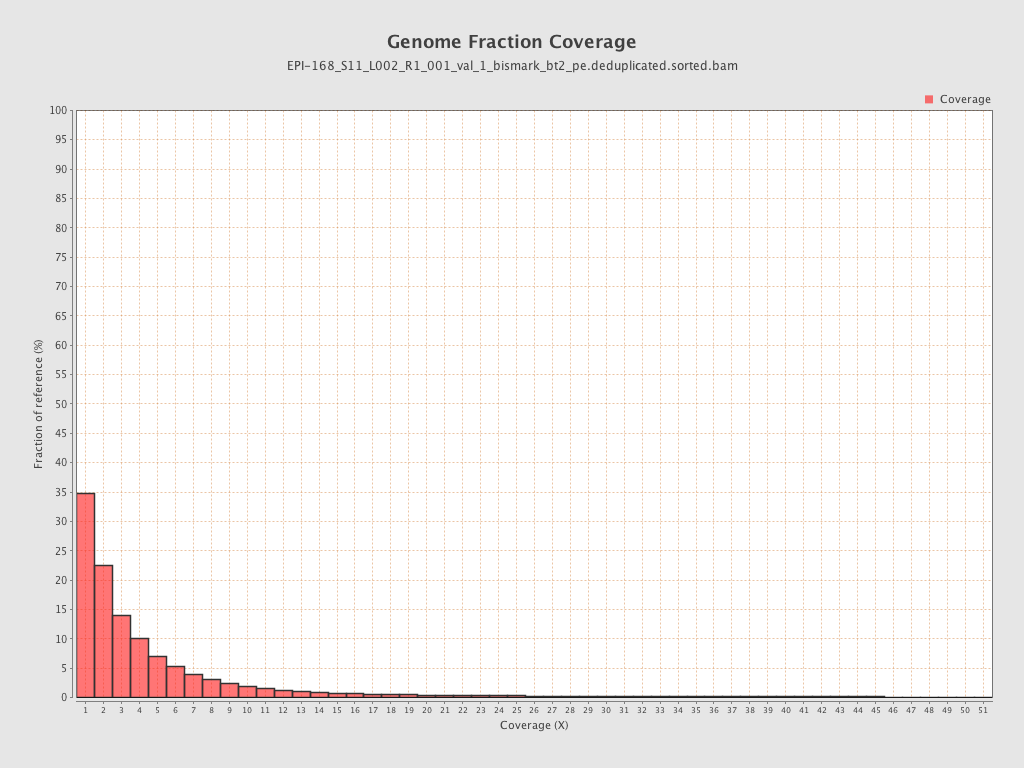
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
117504144 |
1.3108 |
4.7488 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
94523766 |
1.3582 |
6.9248 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
75077322 |
1.3002 |
4.7328 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
80598673 |
1.2345 |
6.8725 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
73165153 |
1.088 |
4.8924 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
73198199 |
1.1852 |
8.8432 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
52966068 |
1.2283 |
4.4356 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
76517797 |
1.2513 |
4.7129 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
54353937 |
1.4088 |
12.3723 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
62470300 |
1.1577 |
3.9196 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
55593706 |
1.0805 |
5.32 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
66462994 |
1.3177 |
6.1062 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
54909335 |
1.2368 |
5.3425 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
73094964 |
1.6103 |
24.2024 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
49629133 |
1.0353 |
4.5172 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
43431691 |
1.358 |
22.046 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
44362830 |
1.2703 |
5.7209 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
31547593 |
1.1374 |
8.4046 |