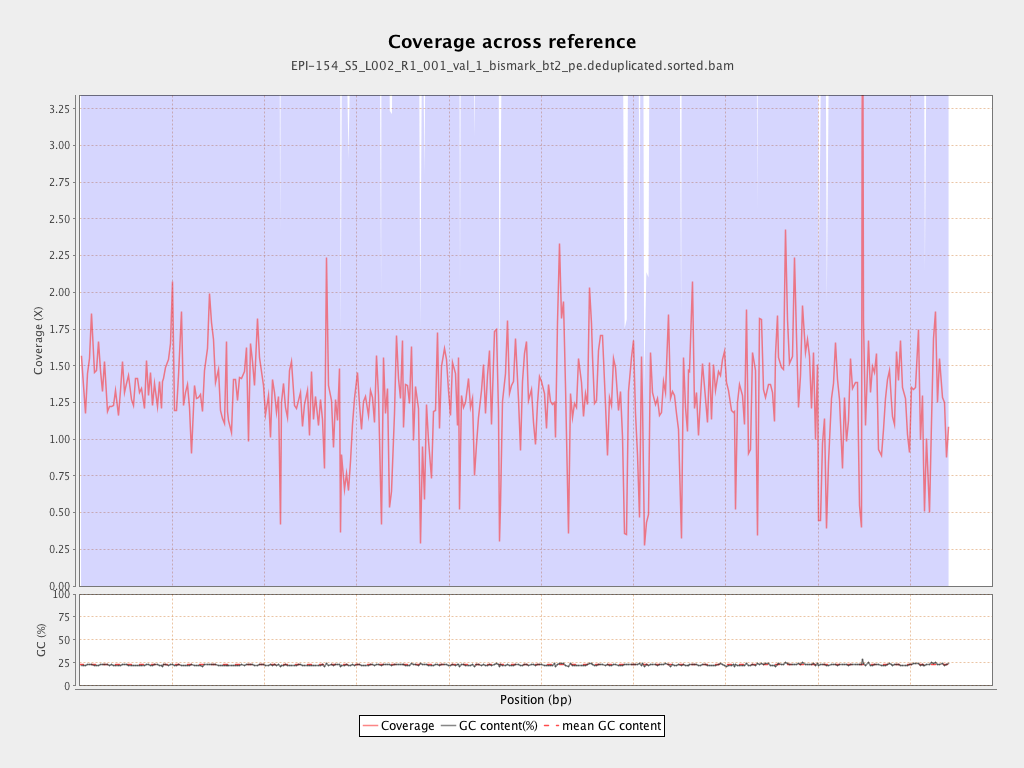
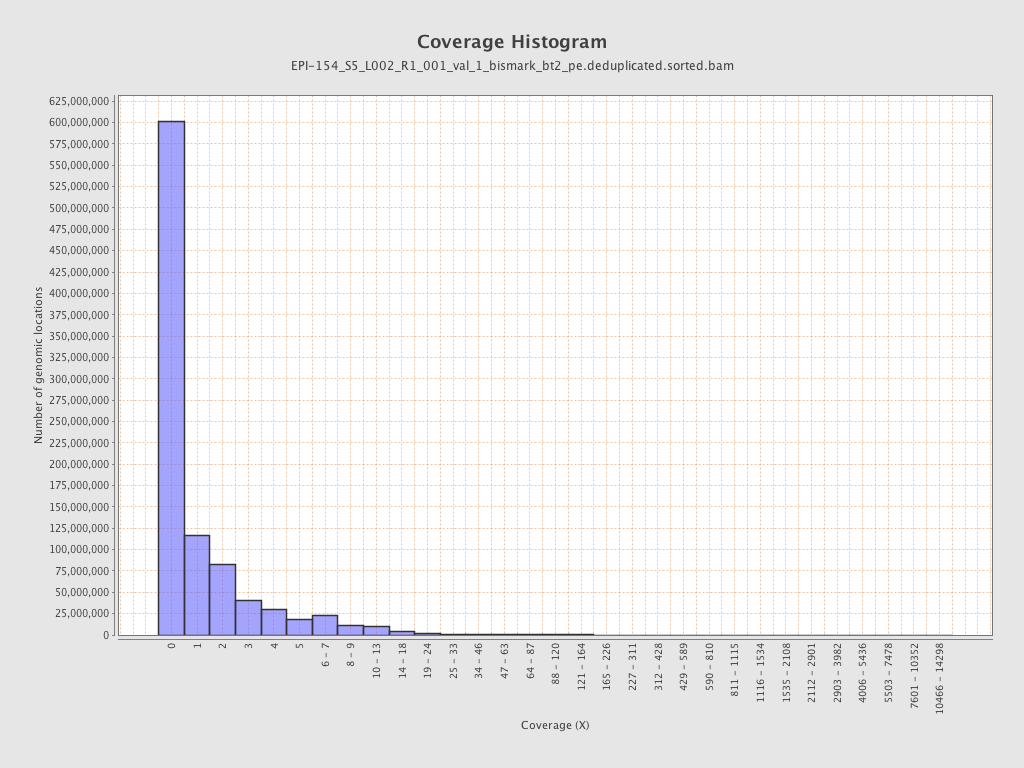
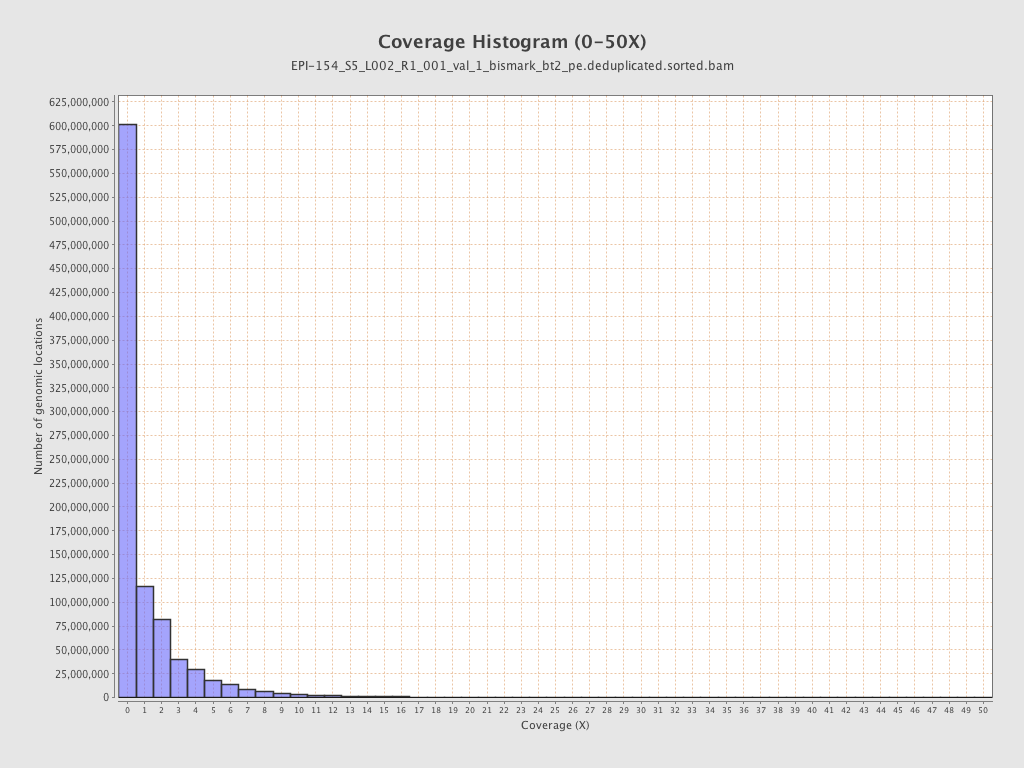
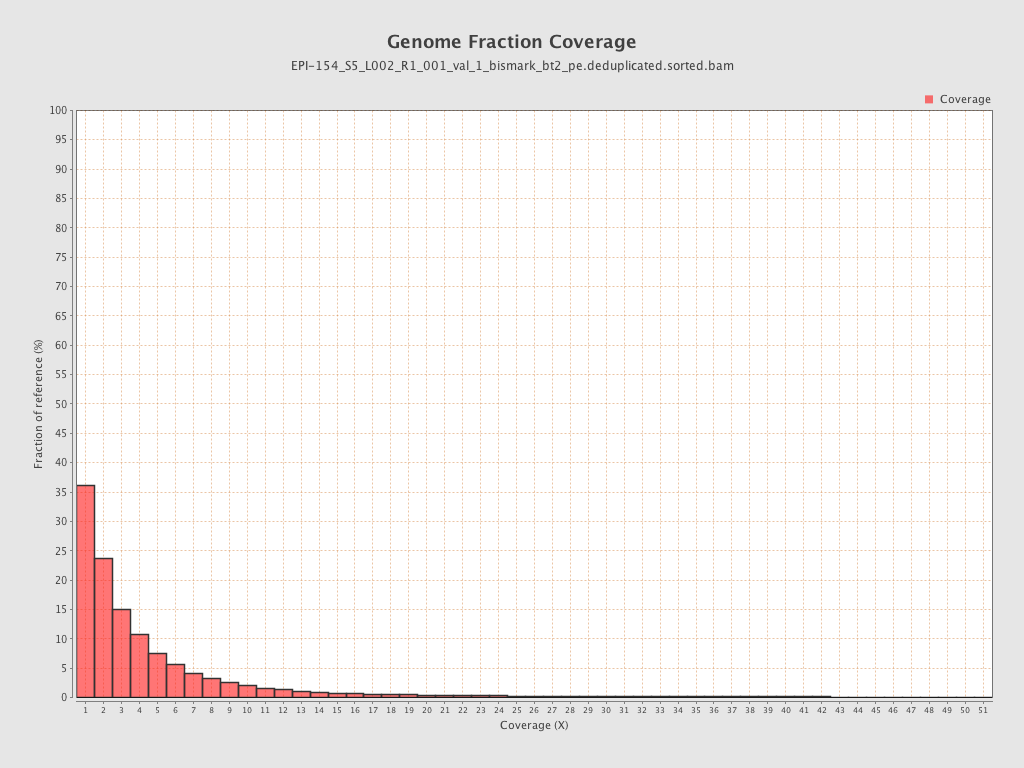
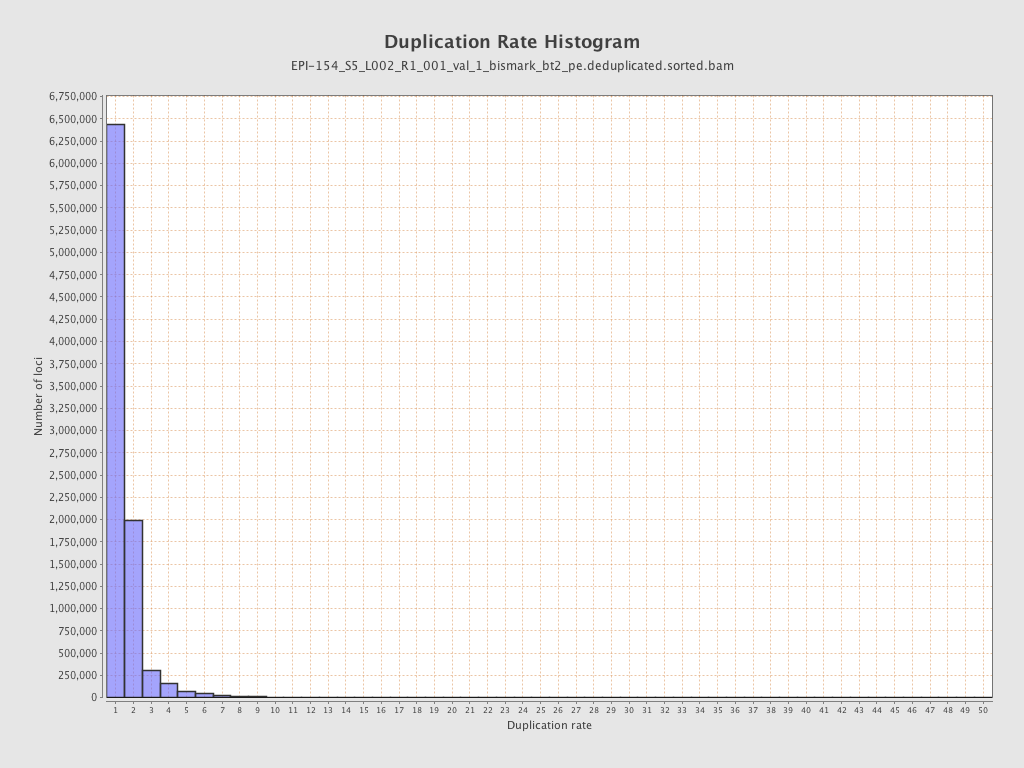
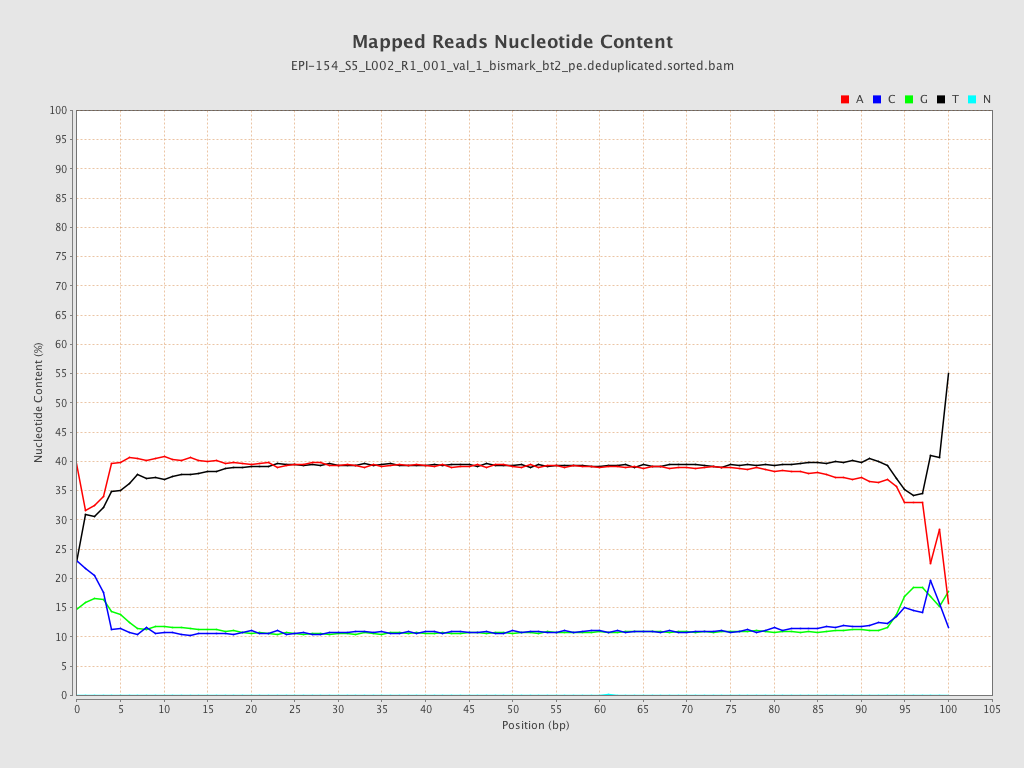
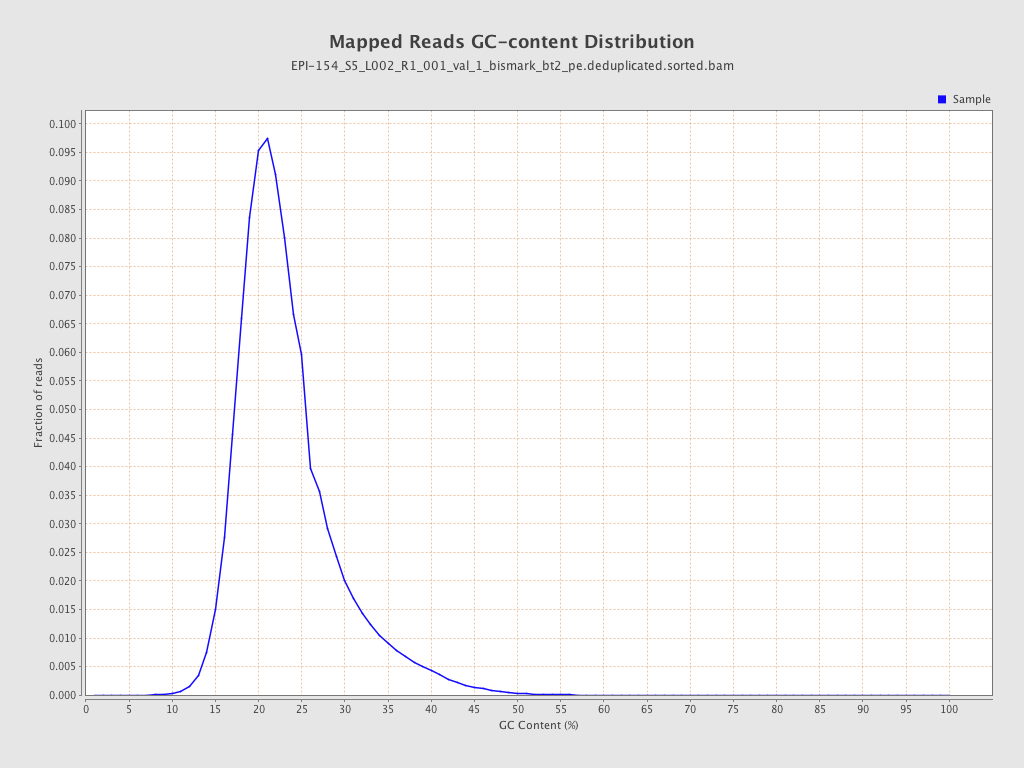
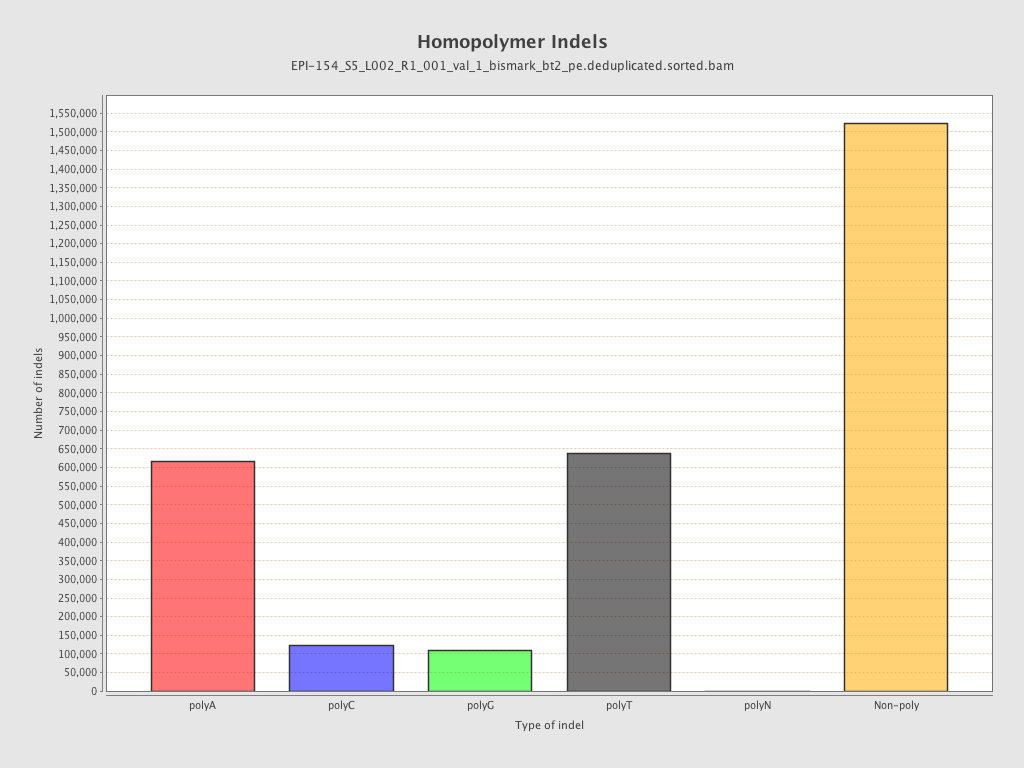
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
122960880 |
1.3717 |
4.1666 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
99152062 |
1.4247 |
6.3795 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
77989152 |
1.3506 |
4.4341 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
82834850 |
1.2688 |
6.5028 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
75756895 |
1.1265 |
4.601 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
75271262 |
1.2188 |
6.422 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
55209059 |
1.2804 |
4.0819 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
79467010 |
1.2995 |
4.6187 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
55927117 |
1.4496 |
11.6885 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
65859743 |
1.2205 |
3.7953 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
58548861 |
1.138 |
5.1332 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
69223995 |
1.3724 |
6.2215 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
57128990 |
1.2868 |
5.0829 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
73373929 |
1.6164 |
22.9405 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
51261006 |
1.0693 |
4.2366 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
44254503 |
1.3838 |
13.6689 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
45463827 |
1.3018 |
4.589 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
32642364 |
1.1768 |
7.4277 |