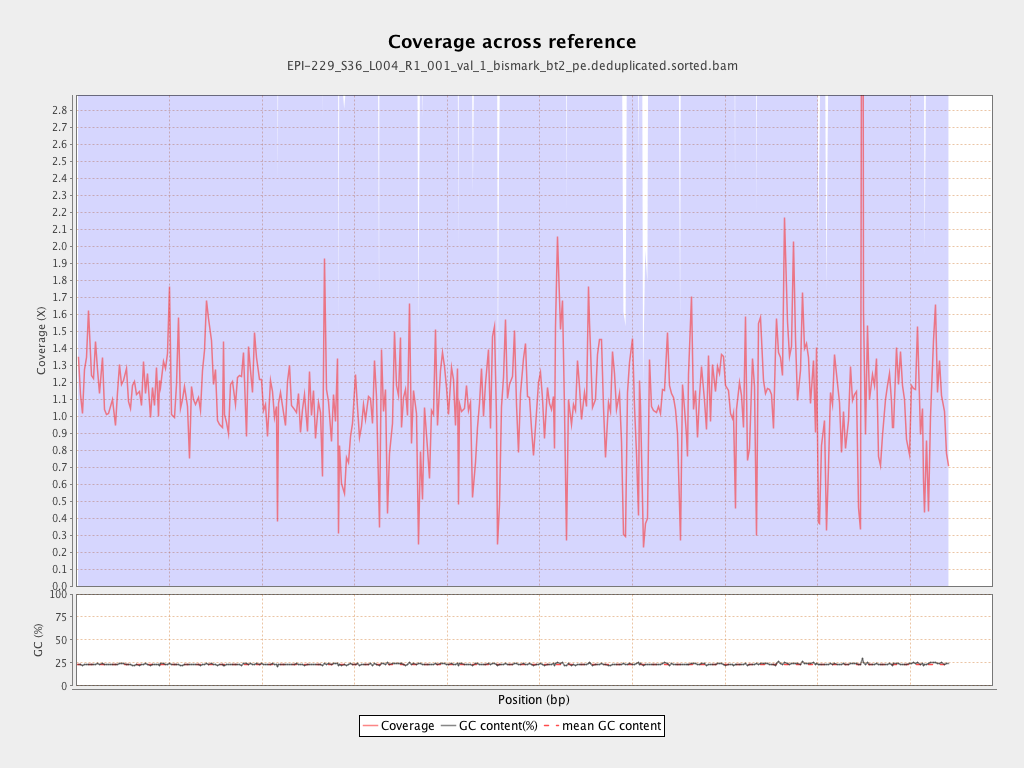
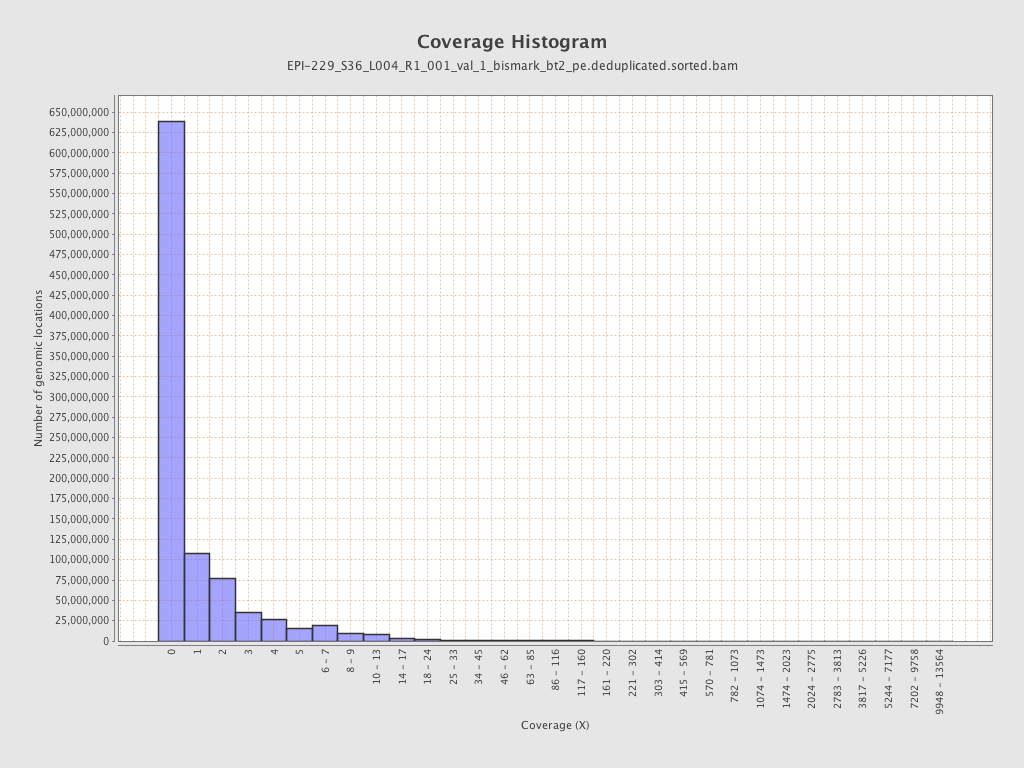
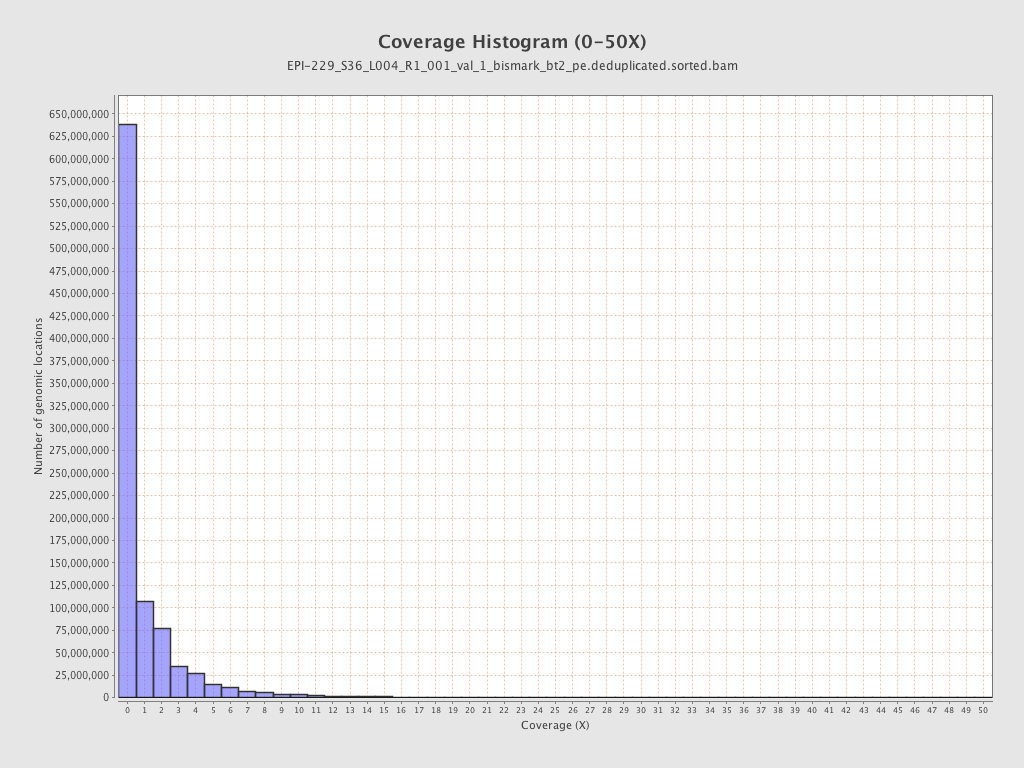
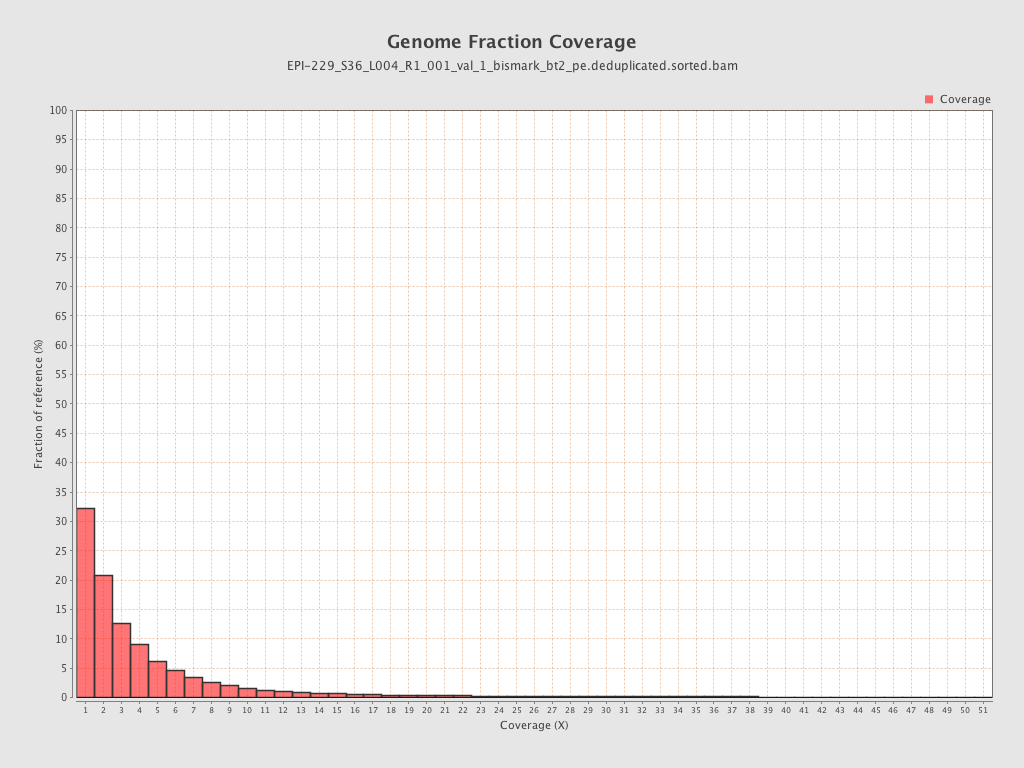
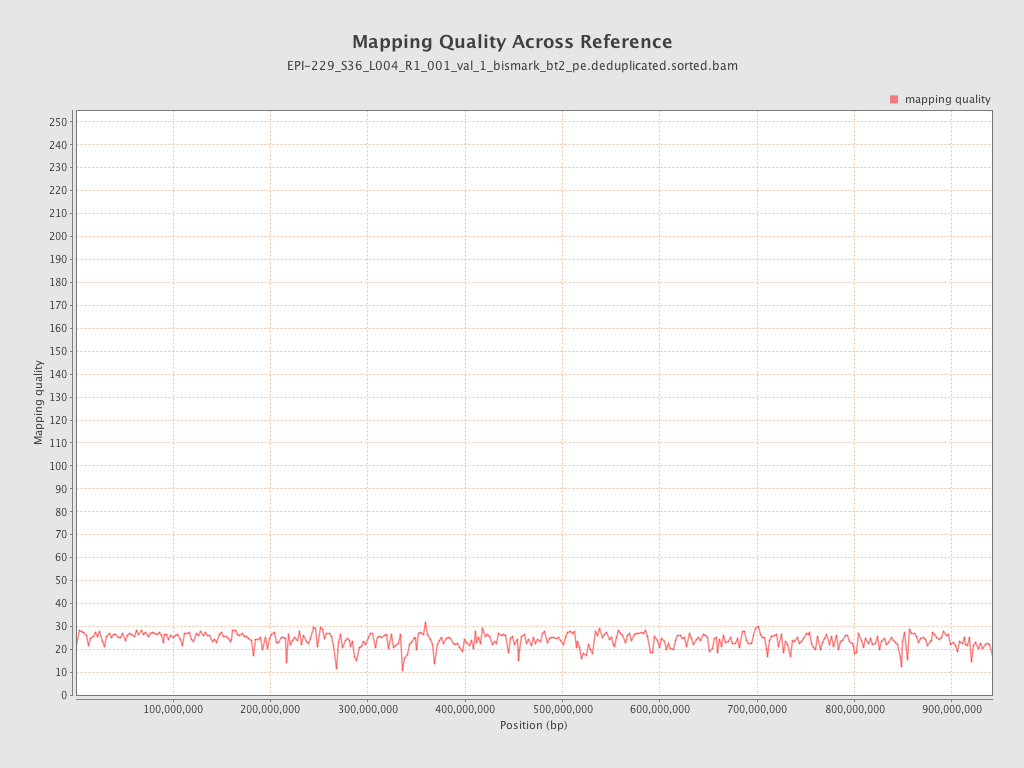
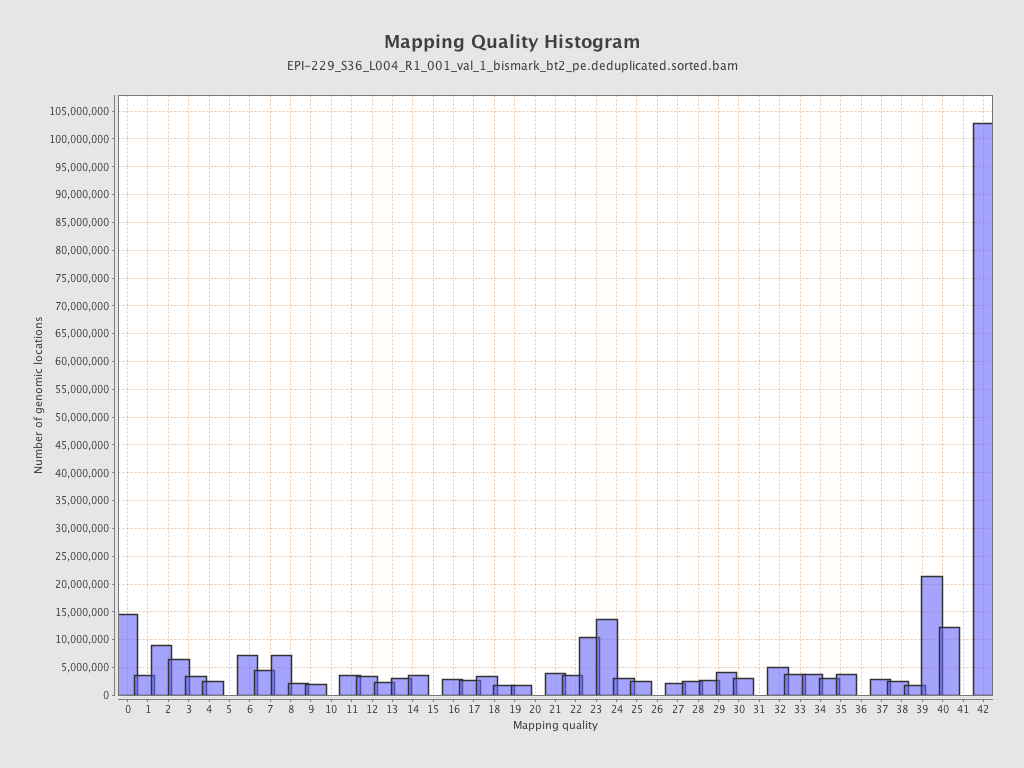
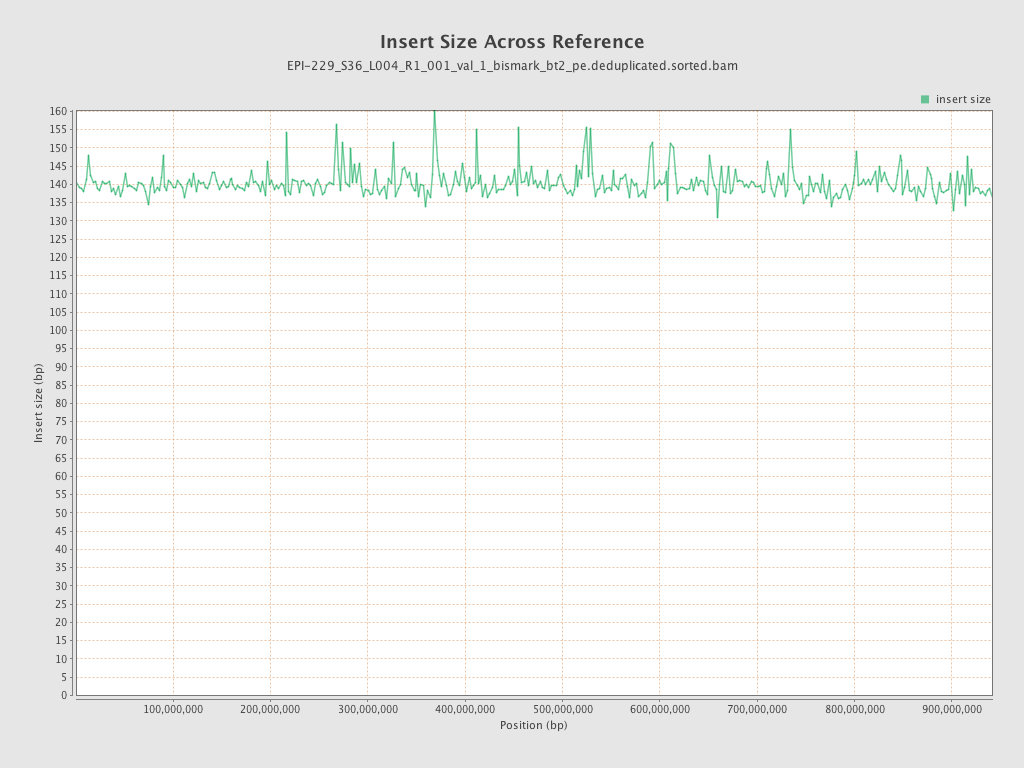
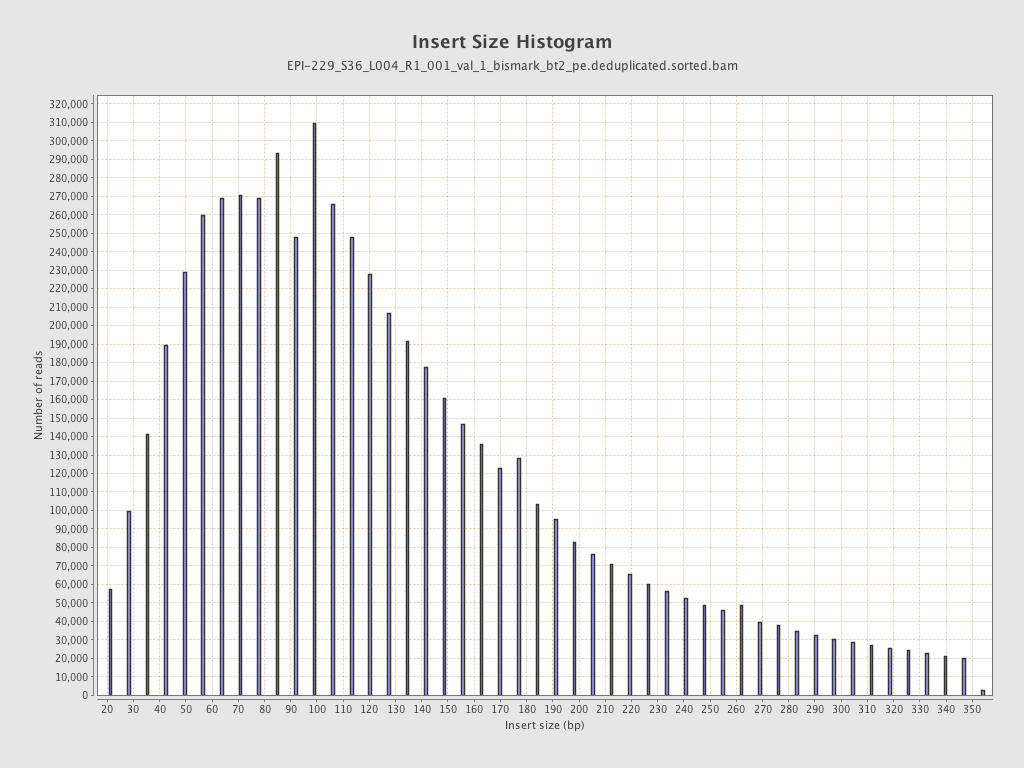
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
105495800 |
1.1768 |
4.0176 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
84180499 |
1.2096 |
5.5811 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
66414636 |
1.1502 |
3.9999 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
70786366 |
1.0842 |
5.822 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
64870036 |
0.9646 |
4.247 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
64425901 |
1.0432 |
7.8409 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
46051439 |
1.068 |
3.6913 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
67667744 |
1.1066 |
4.3129 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
47436423 |
1.2295 |
10.7527 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
56475744 |
1.0466 |
3.4571 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
49350951 |
0.9592 |
4.5073 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
59370342 |
1.1771 |
5.2441 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
48675244 |
1.0964 |
4.5195 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
64529694 |
1.4216 |
21.3809 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
42921783 |
0.8954 |
3.7123 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
38398089 |
1.2007 |
17.8216 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
38816959 |
1.1115 |
4.7287 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
27877237 |
1.005 |
6.7968 |