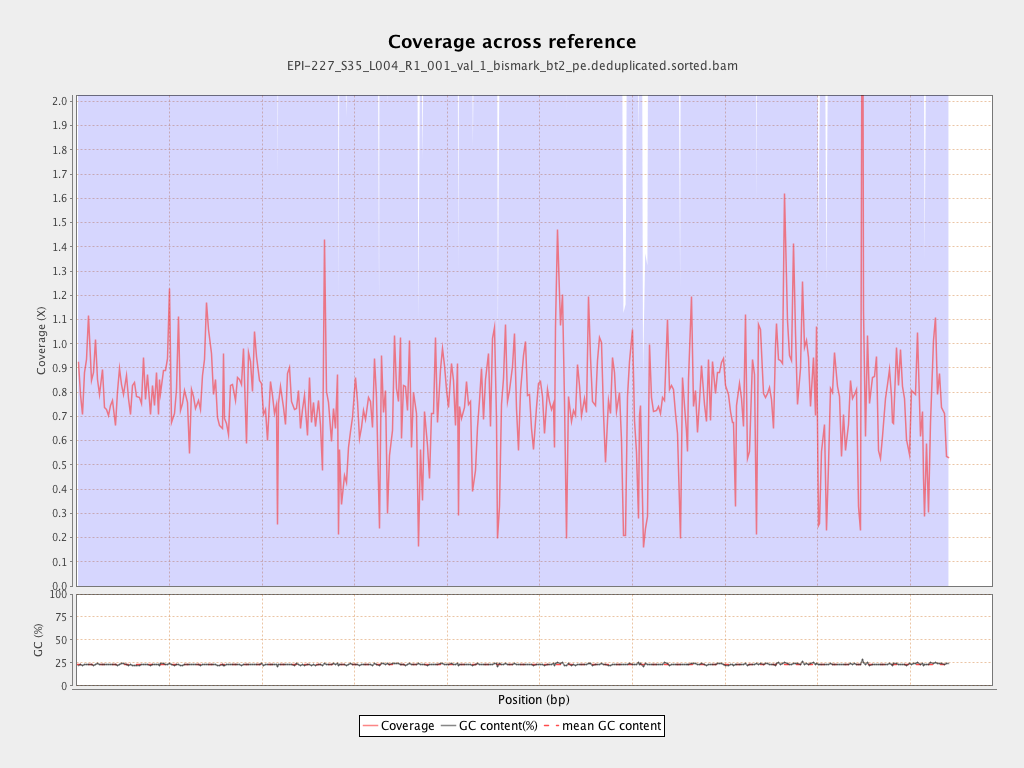
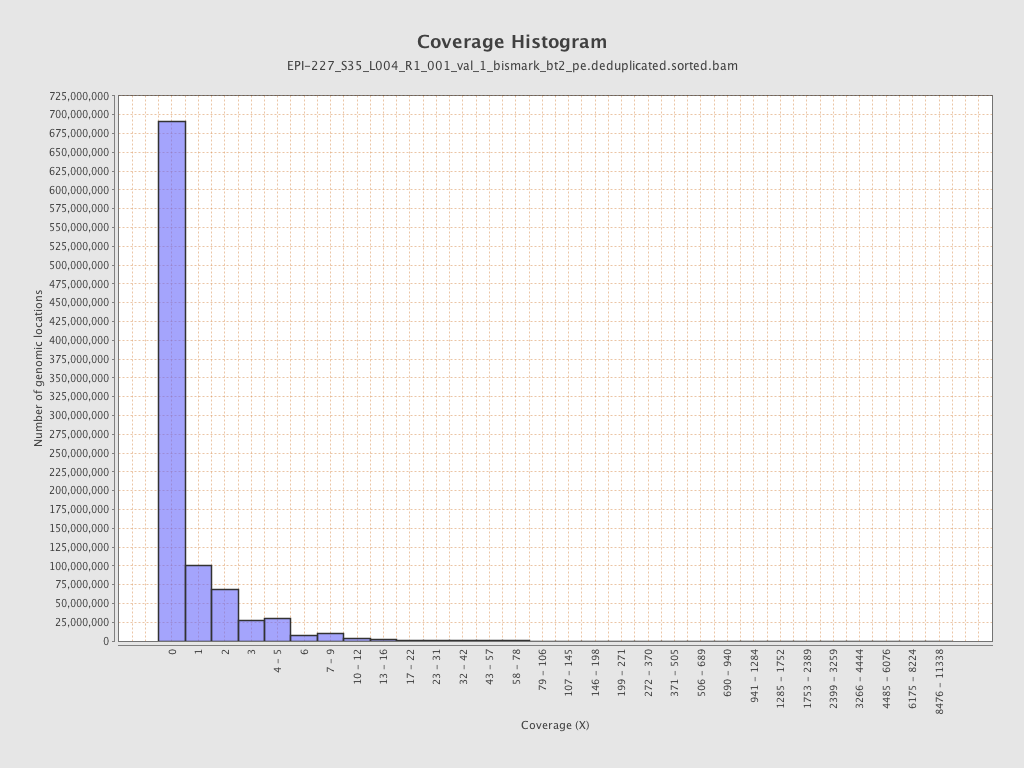
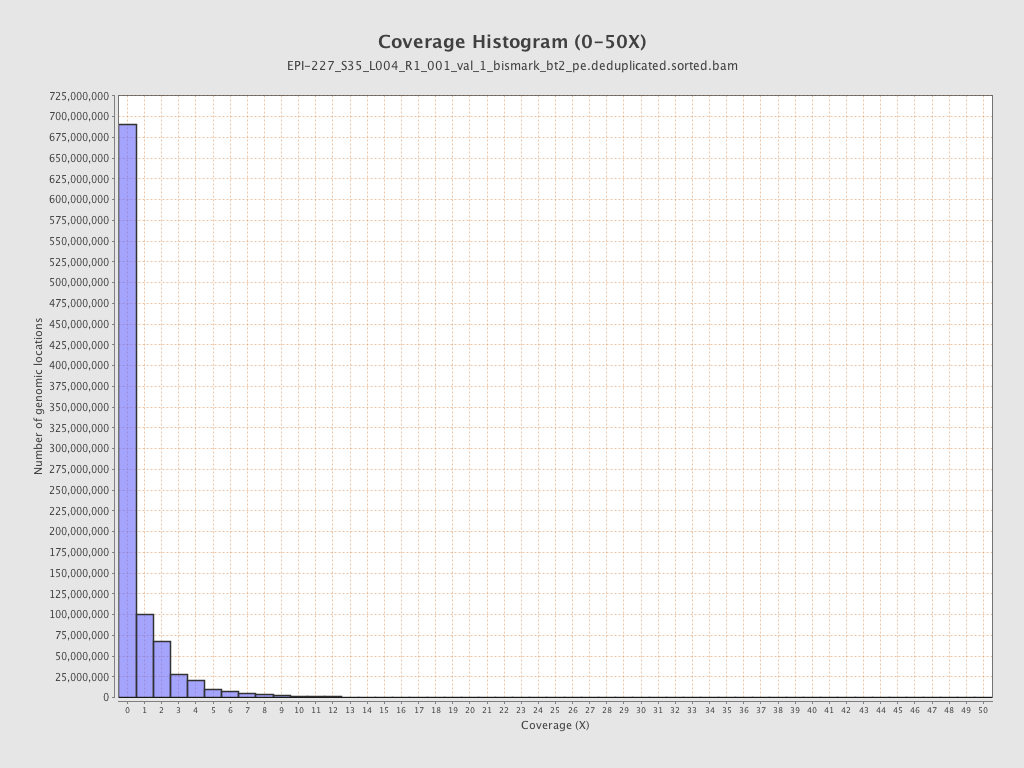
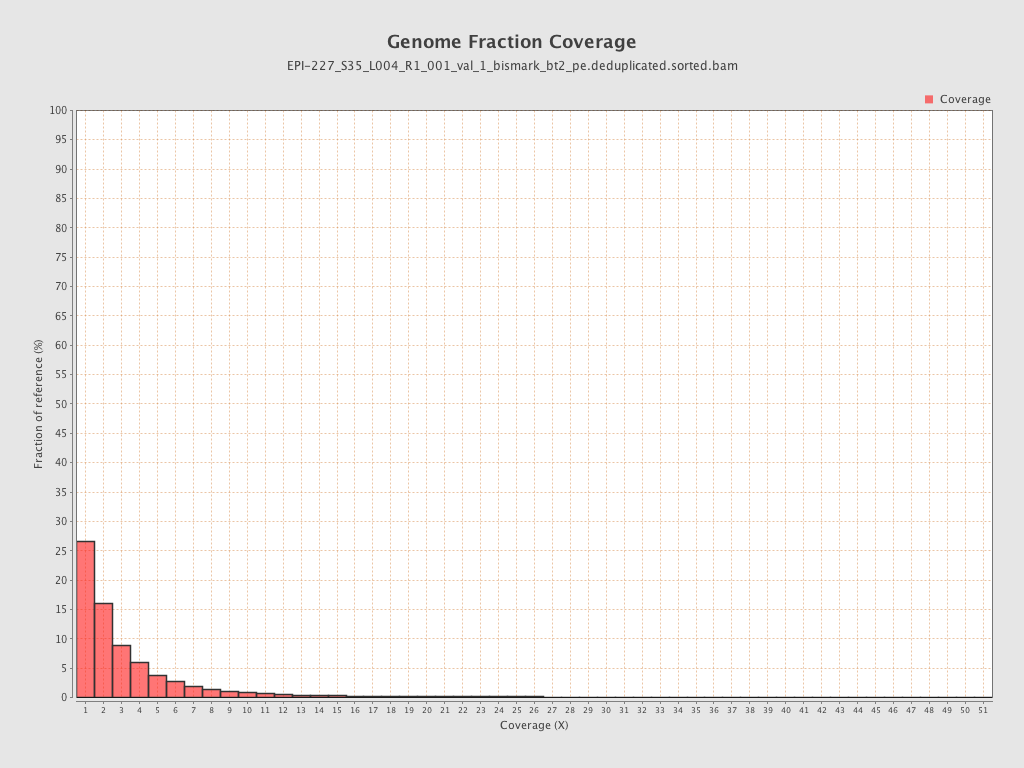
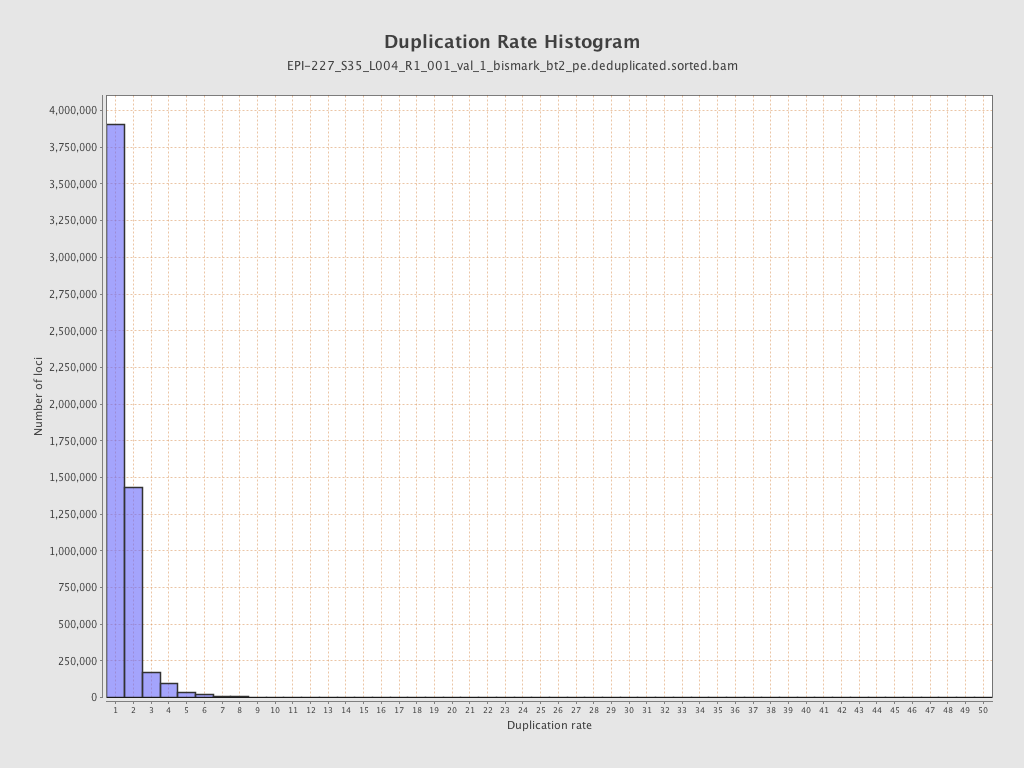
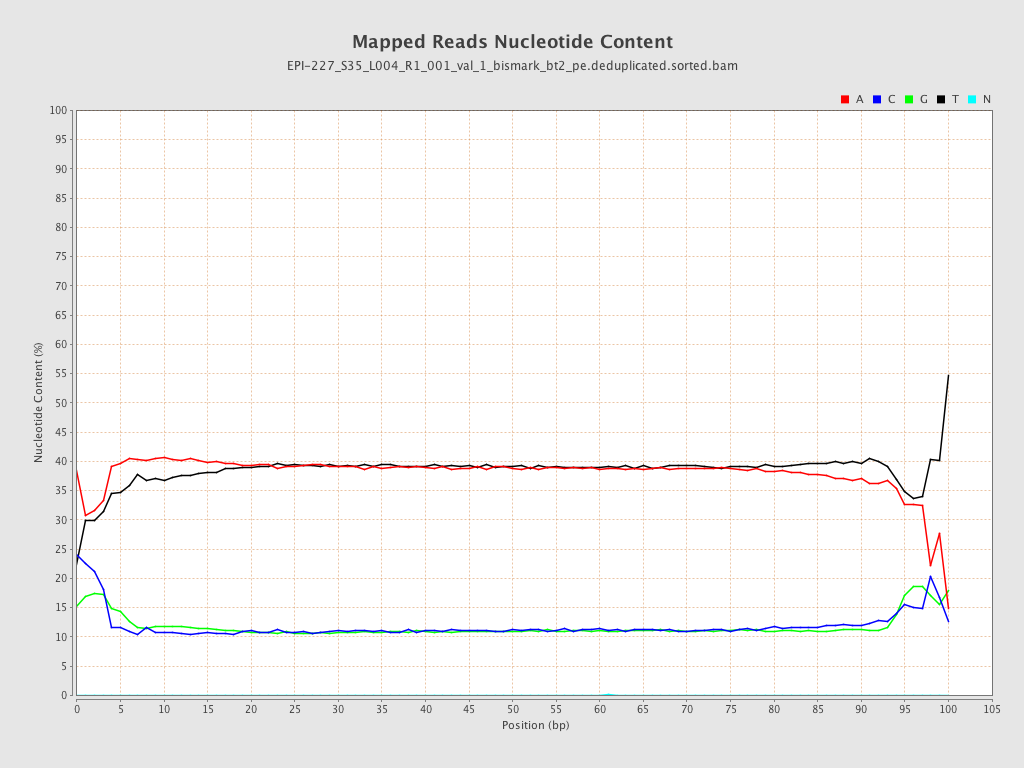
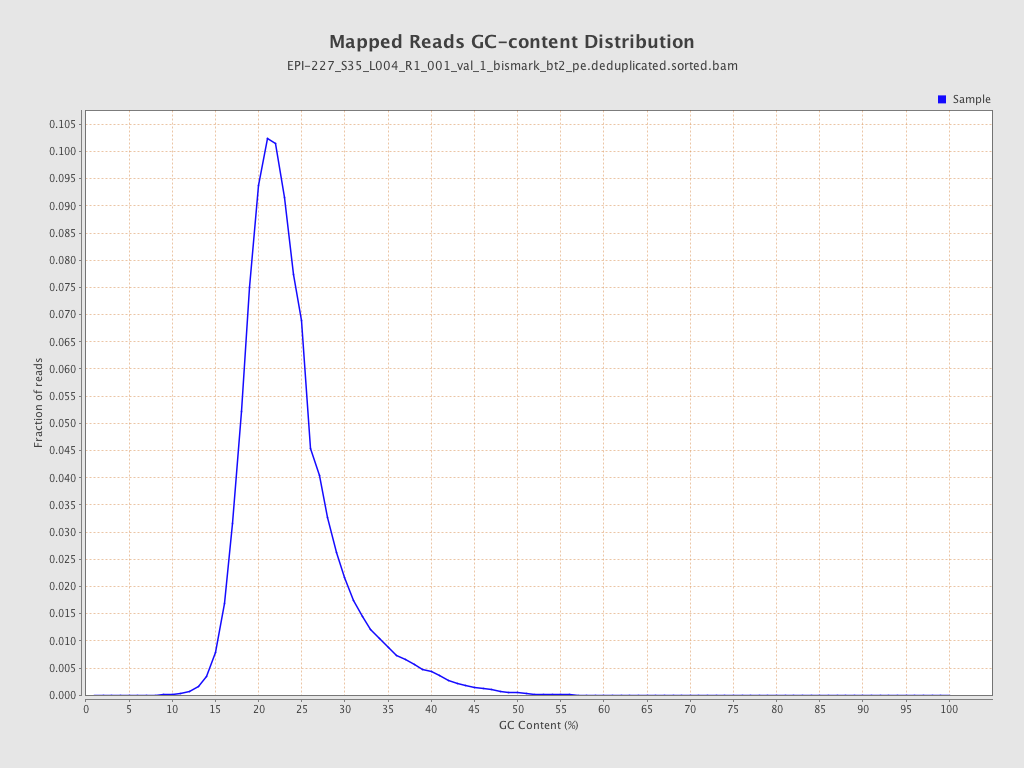
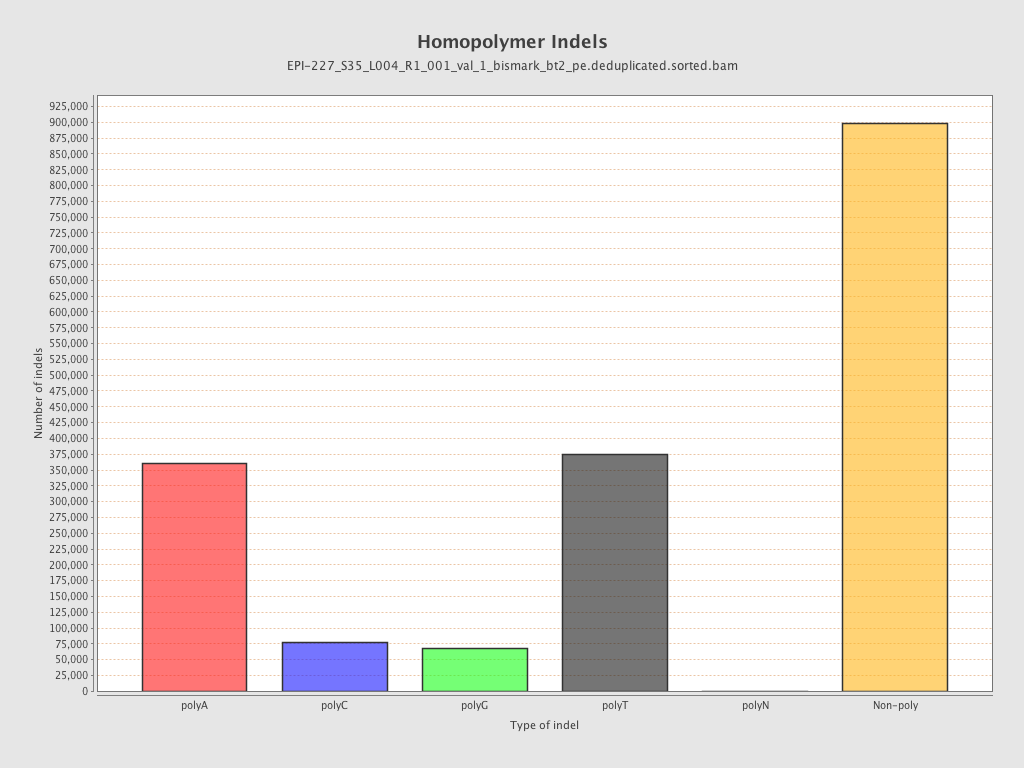
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
73336531 |
0.8181 |
2.6515 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
58064582 |
0.8343 |
3.8965 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
46277468 |
0.8014 |
2.8779 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
49546463 |
0.7589 |
4.3466 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
45002917 |
0.6692 |
2.9736 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
44992123 |
0.7285 |
4.2313 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
32256930 |
0.7481 |
2.583 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
47481491 |
0.7765 |
2.9964 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
33264590 |
0.8622 |
8.1876 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
39185704 |
0.7262 |
2.4642 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
35009654 |
0.6805 |
3.3564 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
41397767 |
0.8208 |
3.8484 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
33914498 |
0.7639 |
3.1163 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
45799938 |
1.009 |
17.2974 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
29981520 |
0.6254 |
2.671 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
26557154 |
0.8304 |
9.3966 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
27052361 |
0.7746 |
2.9658 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
19045793 |
0.6866 |
4.521 |