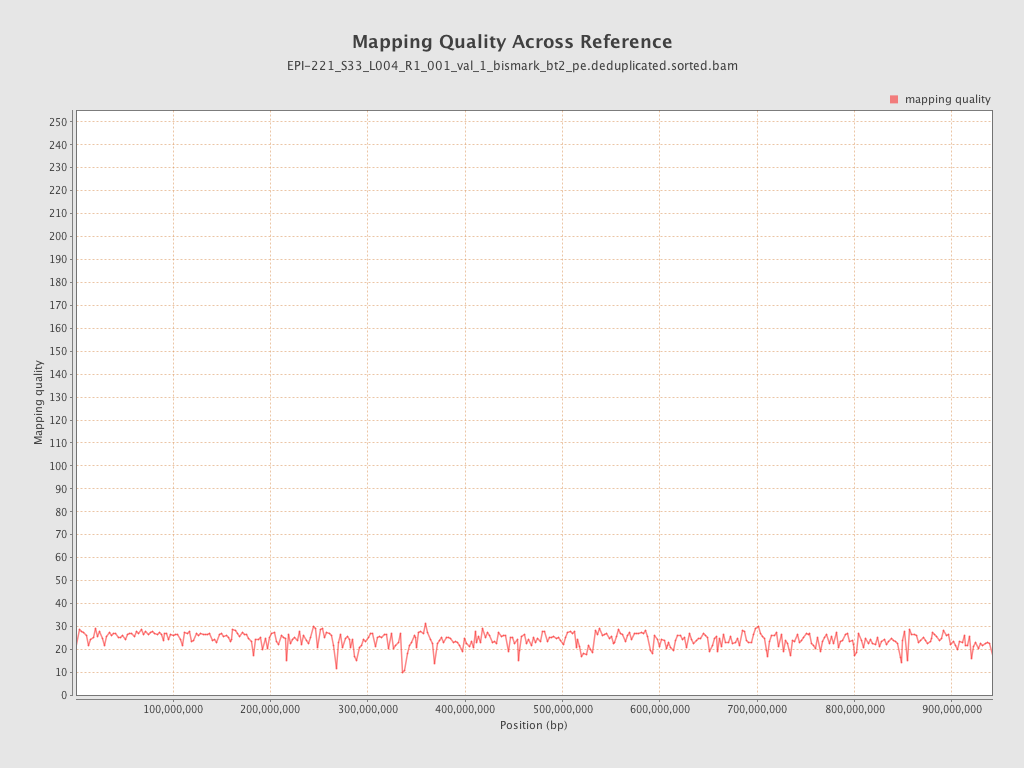
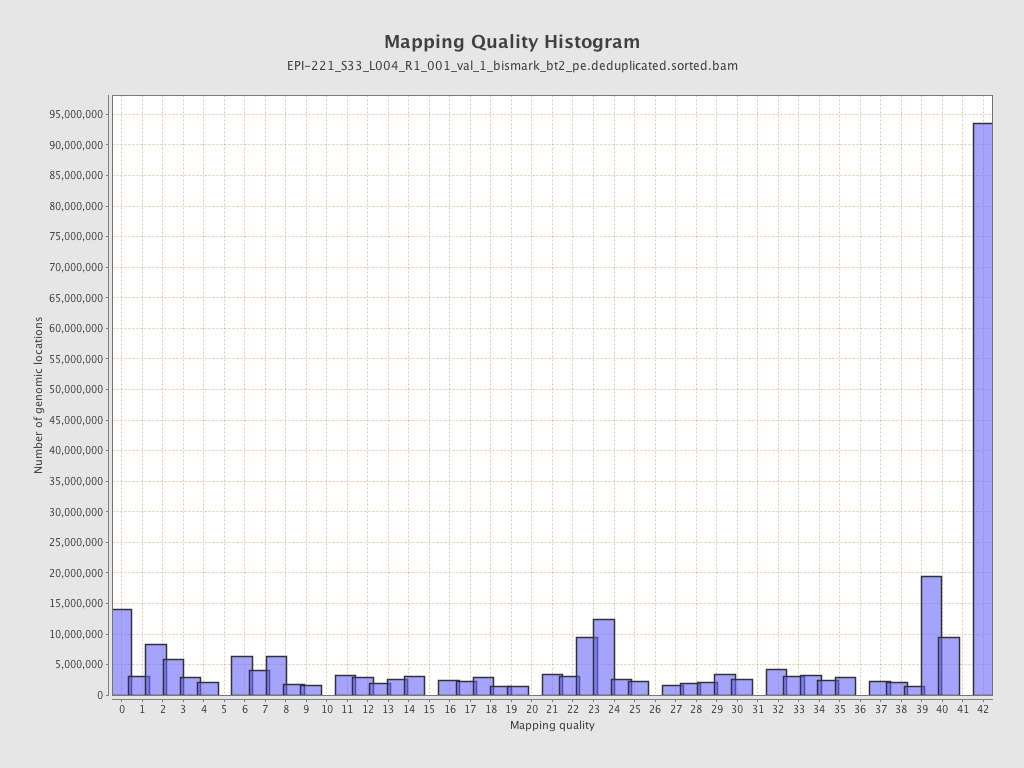
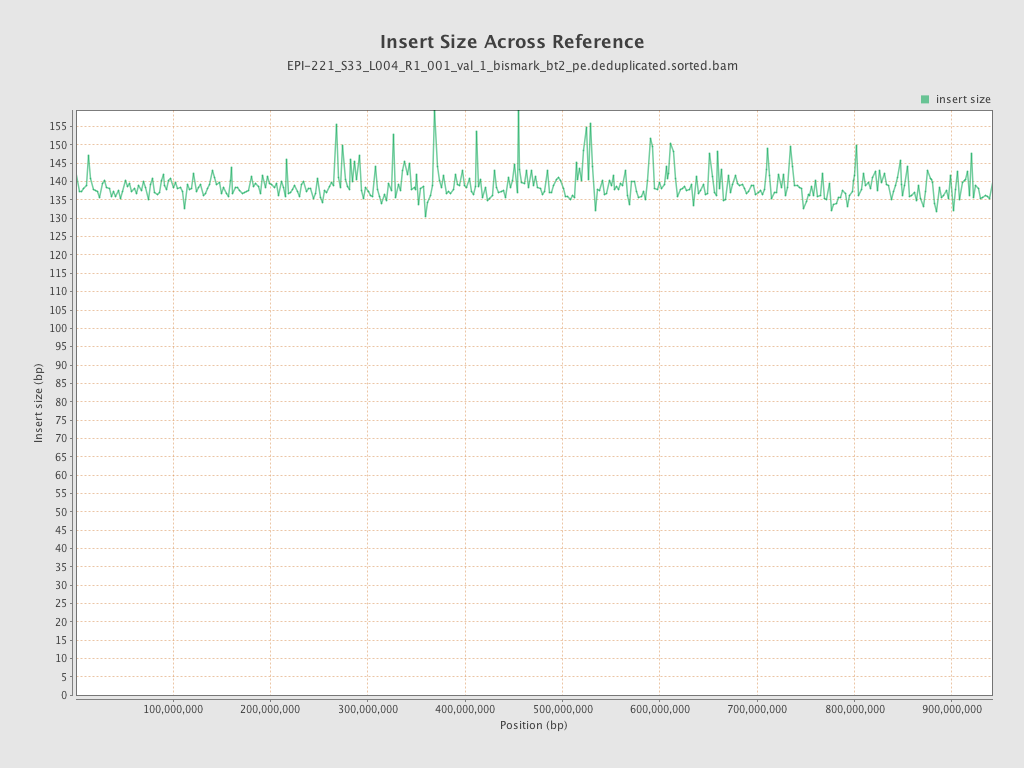
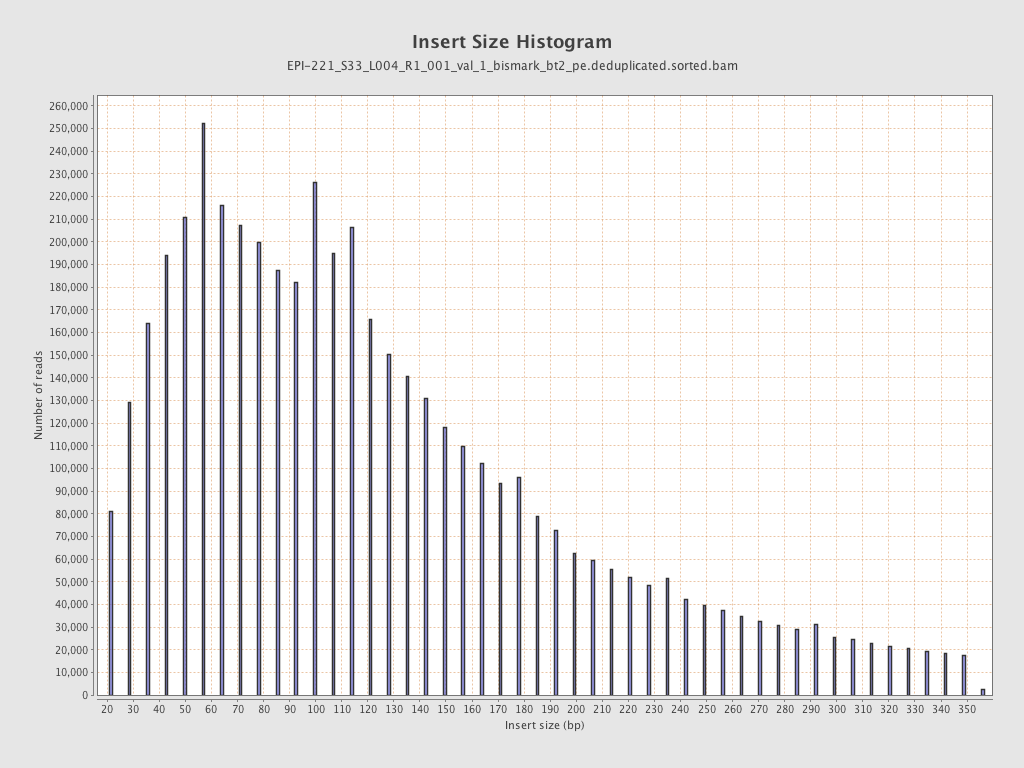
| Name |
Length |
Mapped bases |
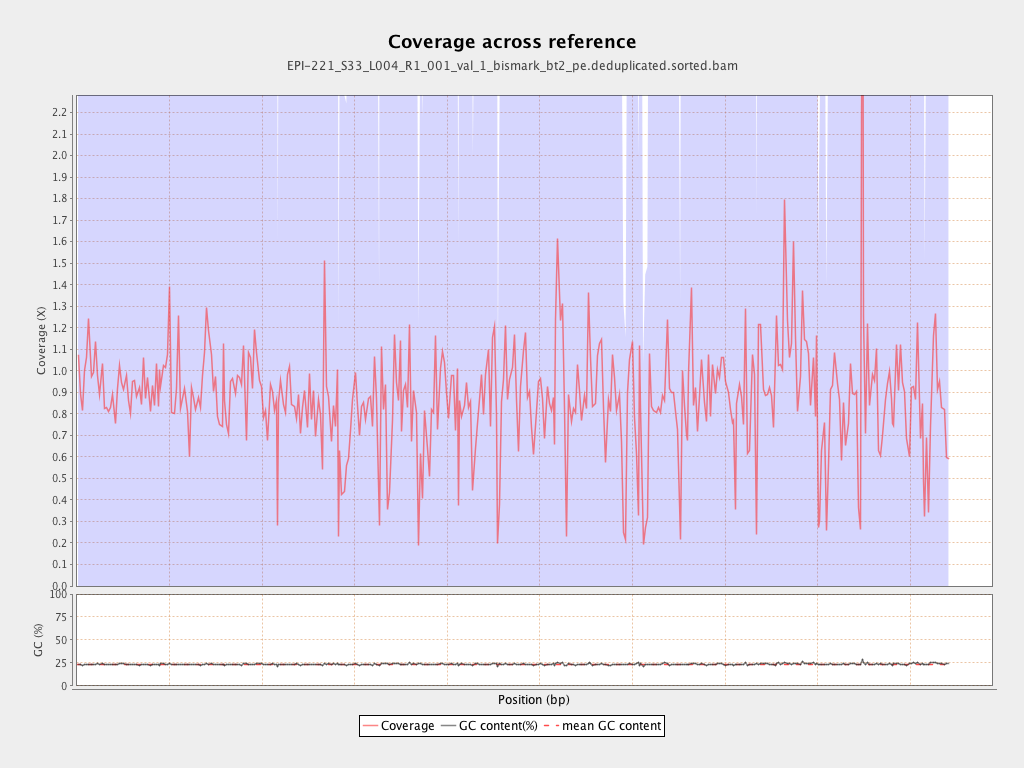
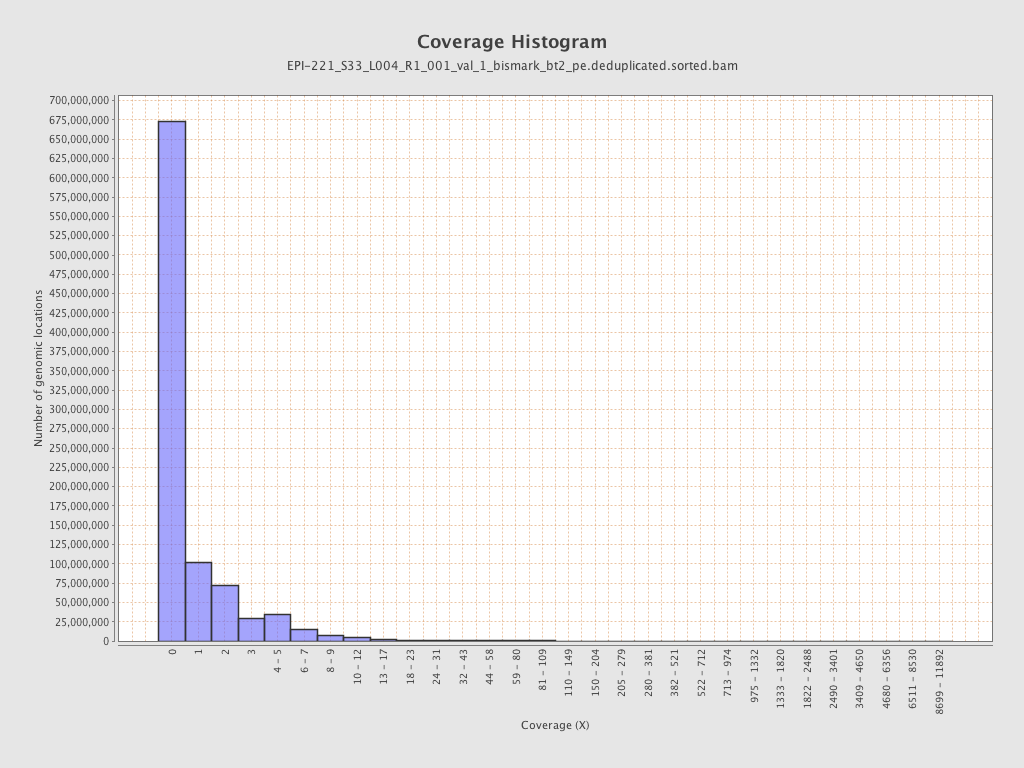
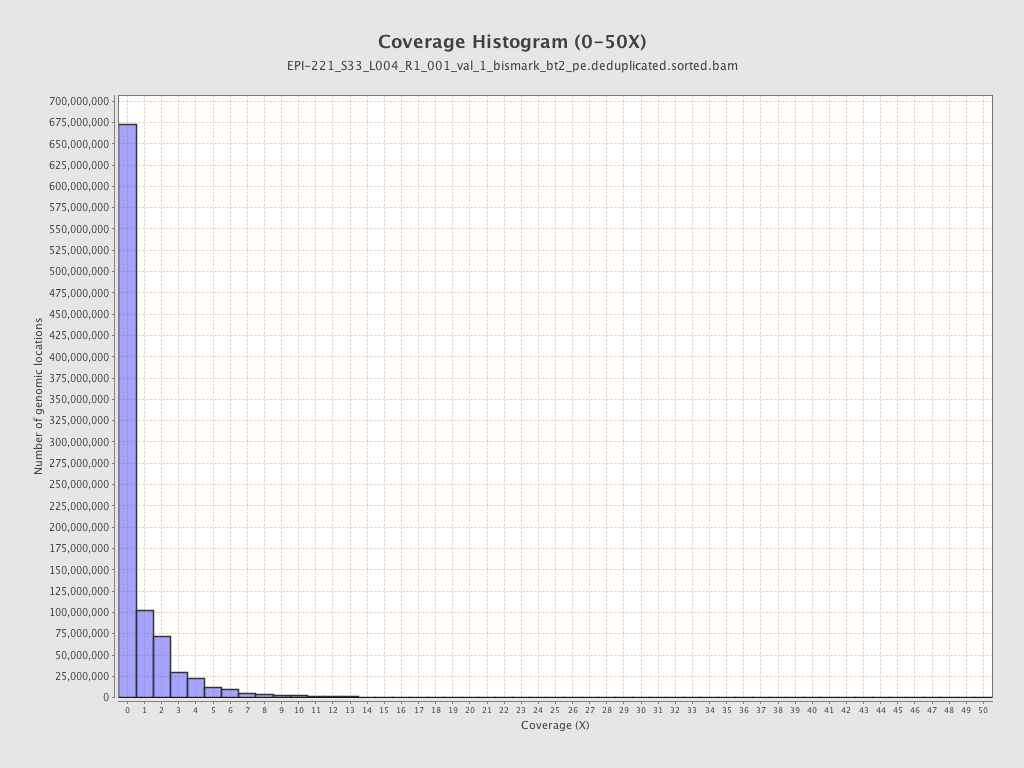
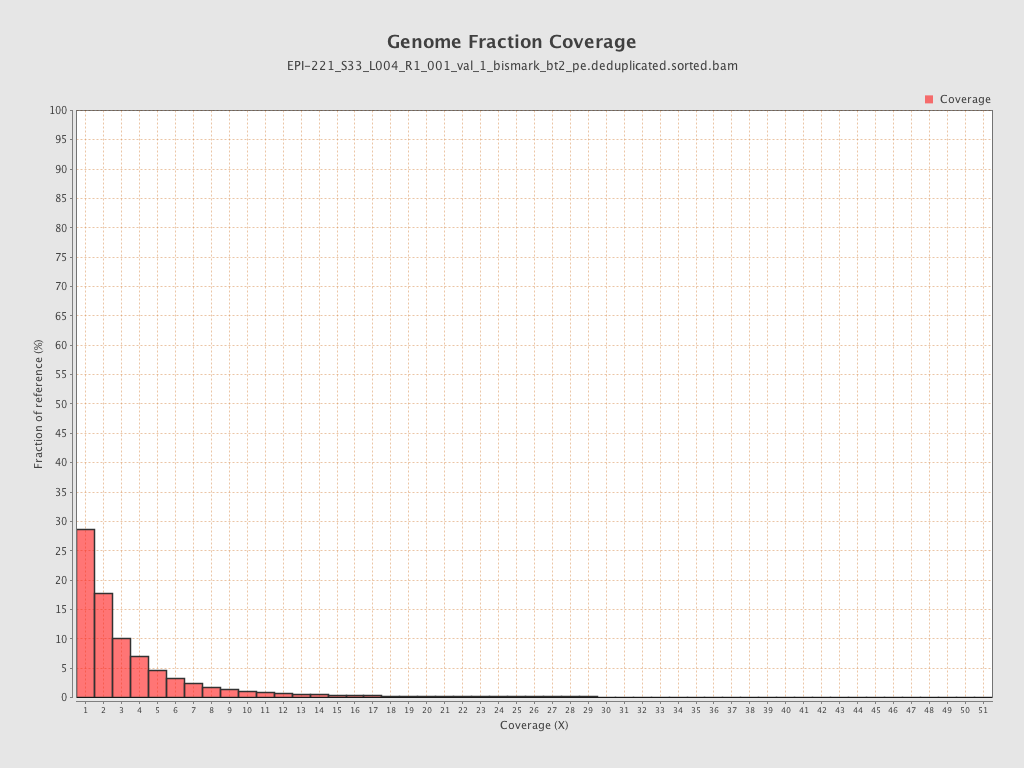
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
83444154 |
0.9308 |
2.9944 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
65892338 |
0.9468 |
4.3847 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
52599476 |
0.9109 |
3.1701 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
56245205 |
0.8615 |
4.5432 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
51371126 |
0.7639 |
3.3749 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
50642314 |
0.82 |
5.372 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
36849758 |
0.8546 |
2.9765 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
53865828 |
0.8809 |
3.5018 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
37445402 |
0.9705 |
8.8095 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
44235294 |
0.8198 |
2.6886 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
39249561 |
0.7629 |
3.7318 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
47189554 |
0.9356 |
4.3309 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
38527219 |
0.8678 |
3.6068 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
51348414 |
1.1312 |
18.0771 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
34237901 |
0.7142 |
2.9345 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
30243227 |
0.9457 |
11.5118 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
30793575 |
0.8817 |
3.427 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
21574253 |
0.7778 |
5.0117 |