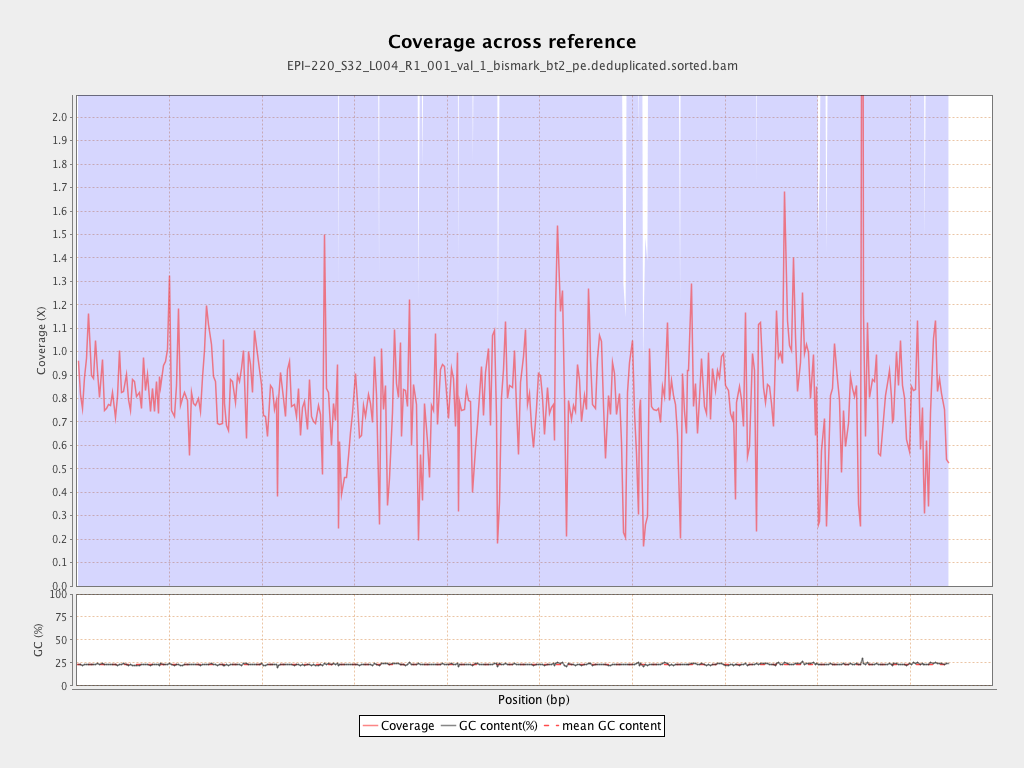
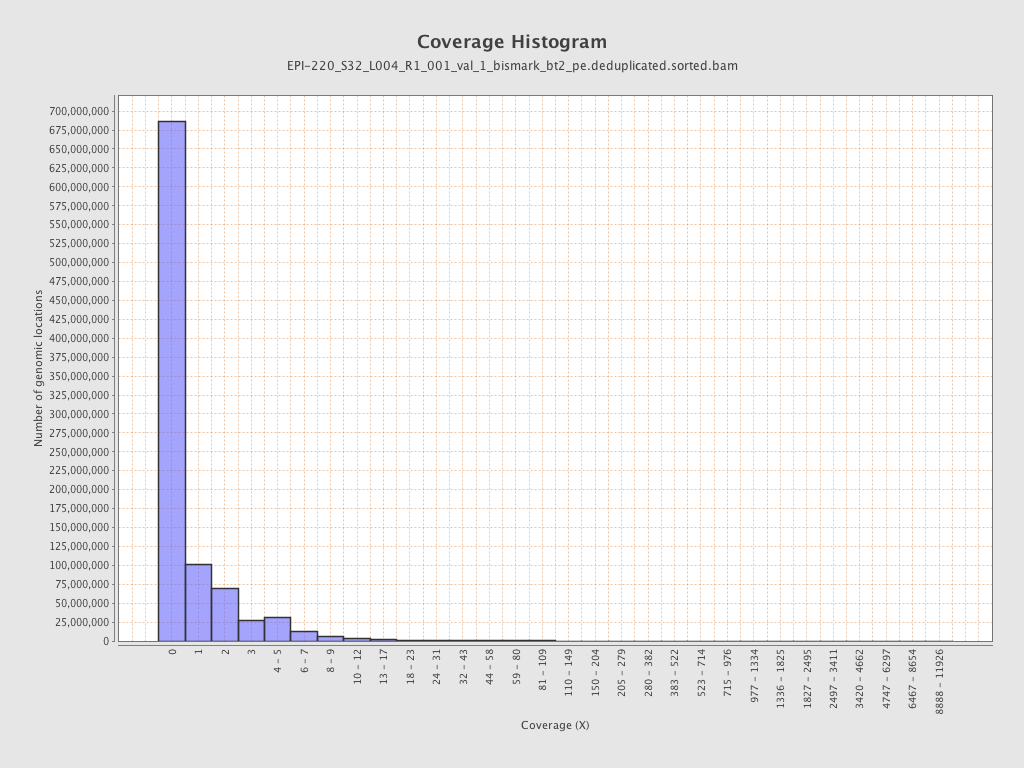
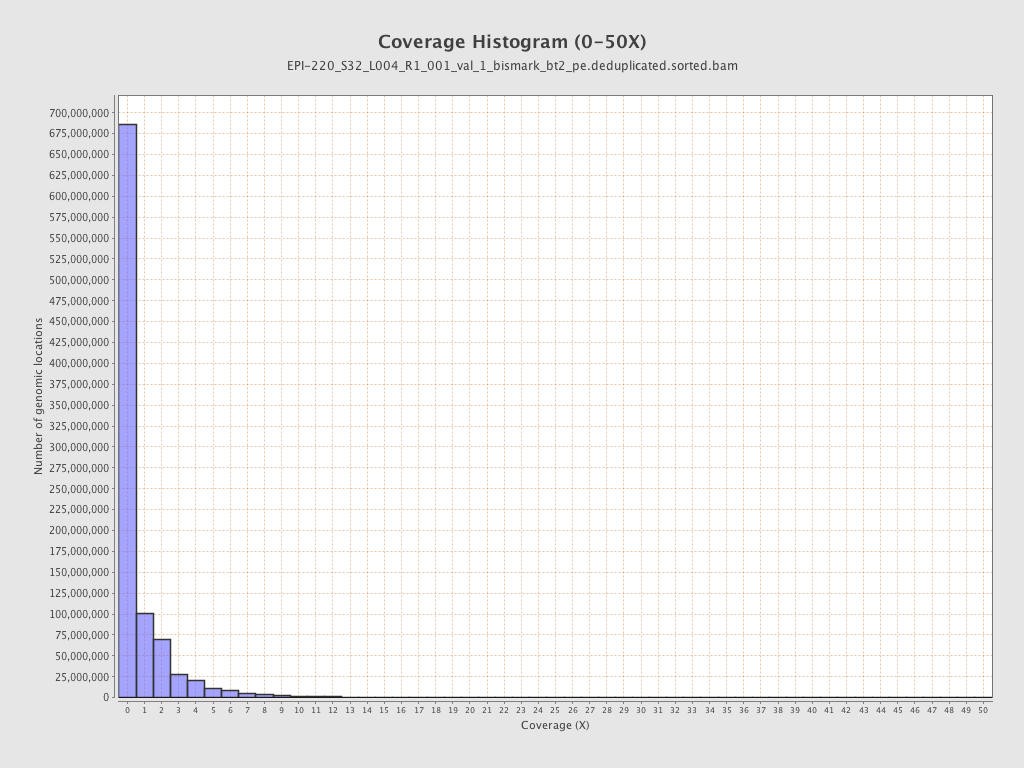
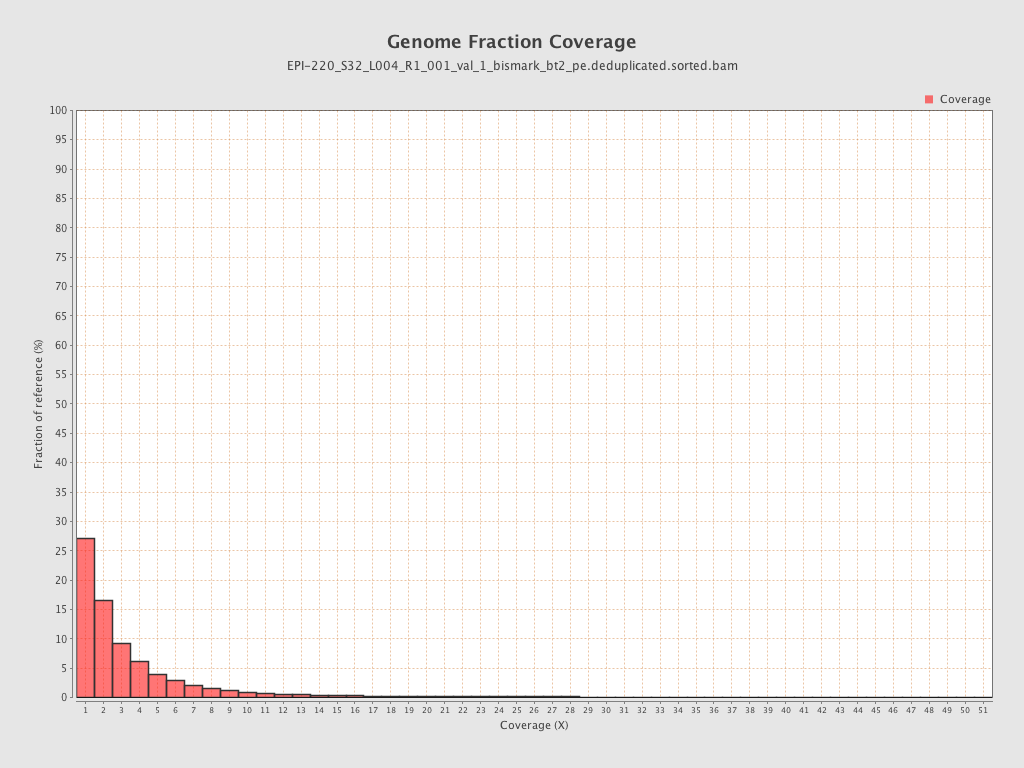
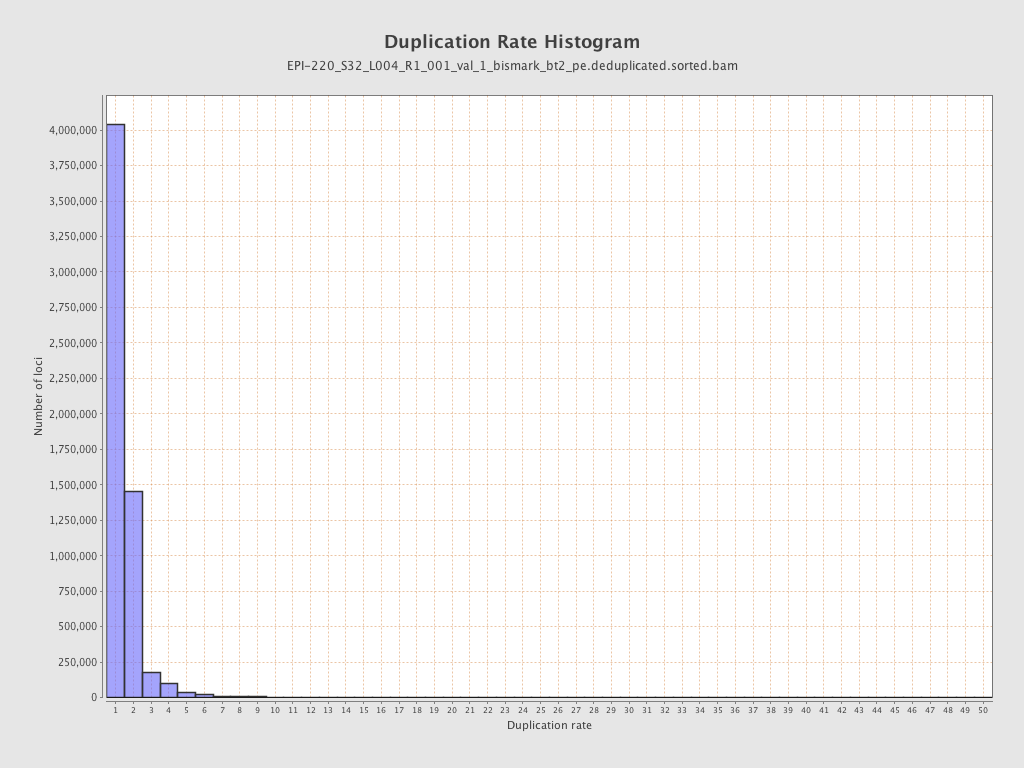
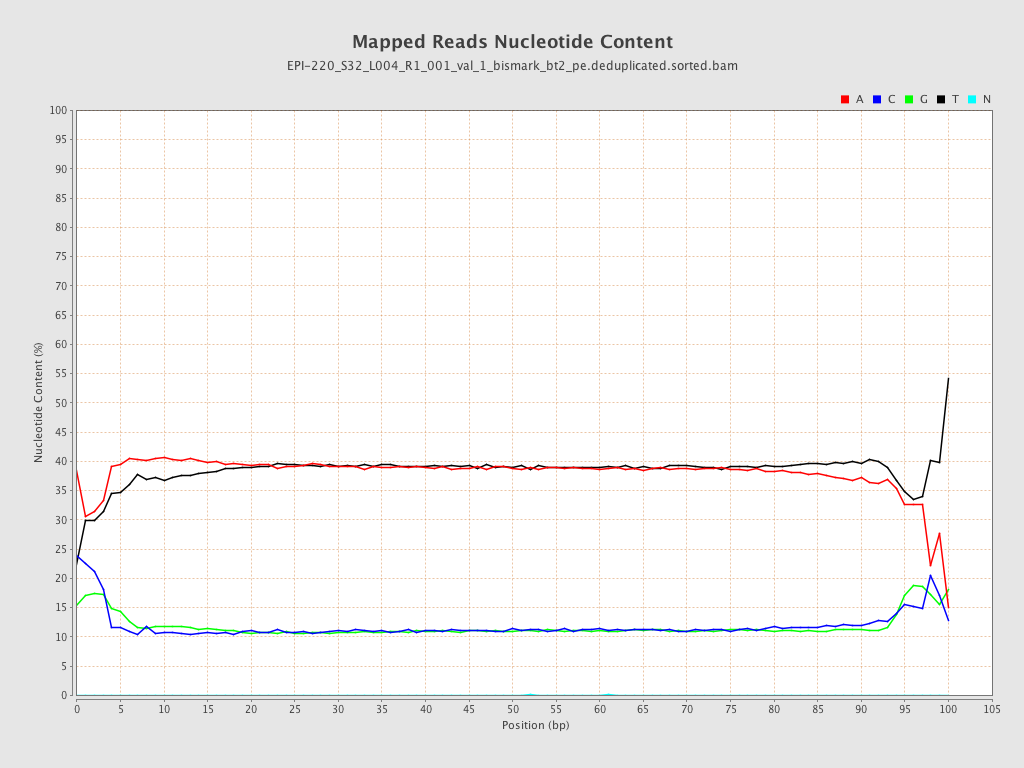
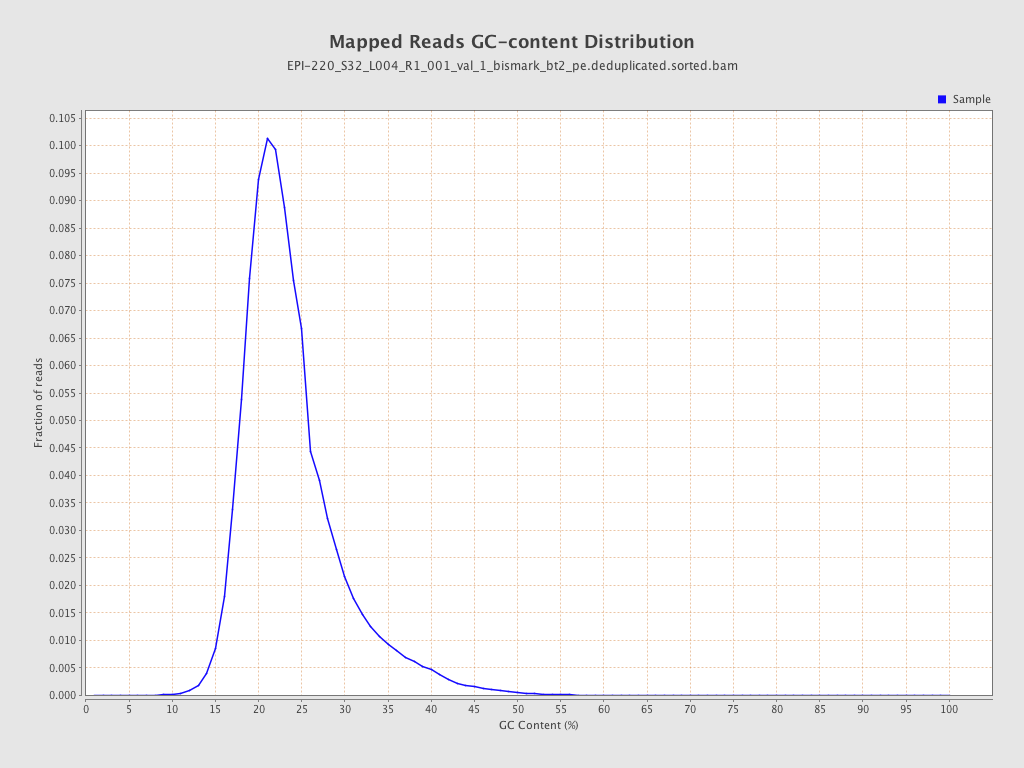
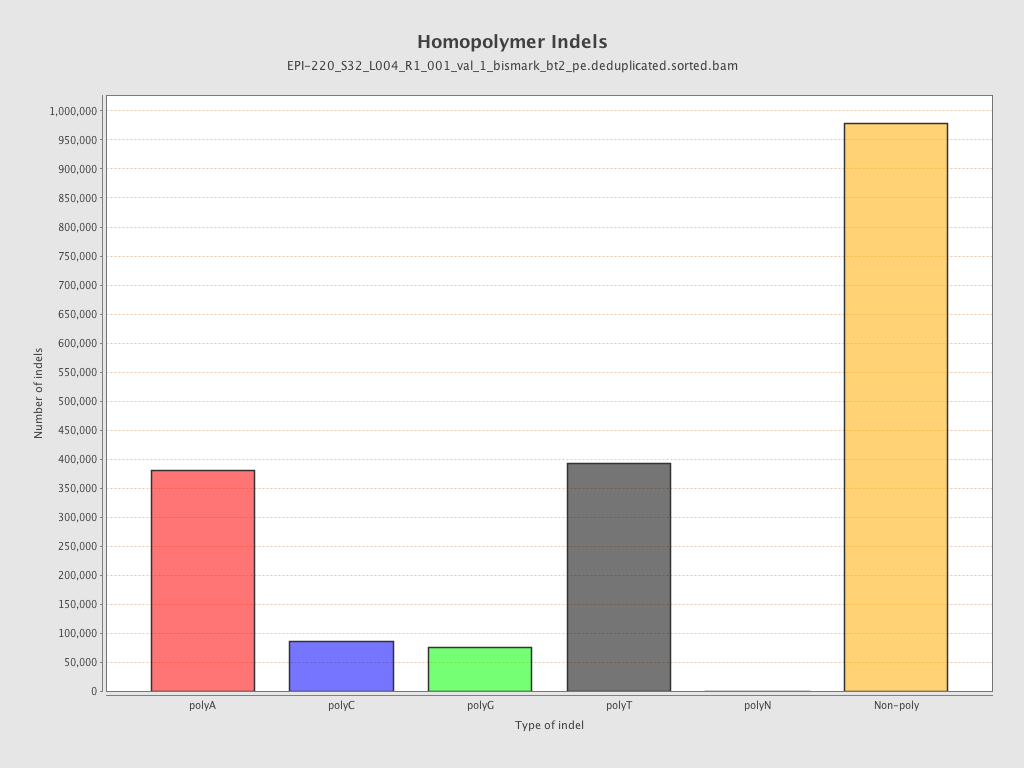
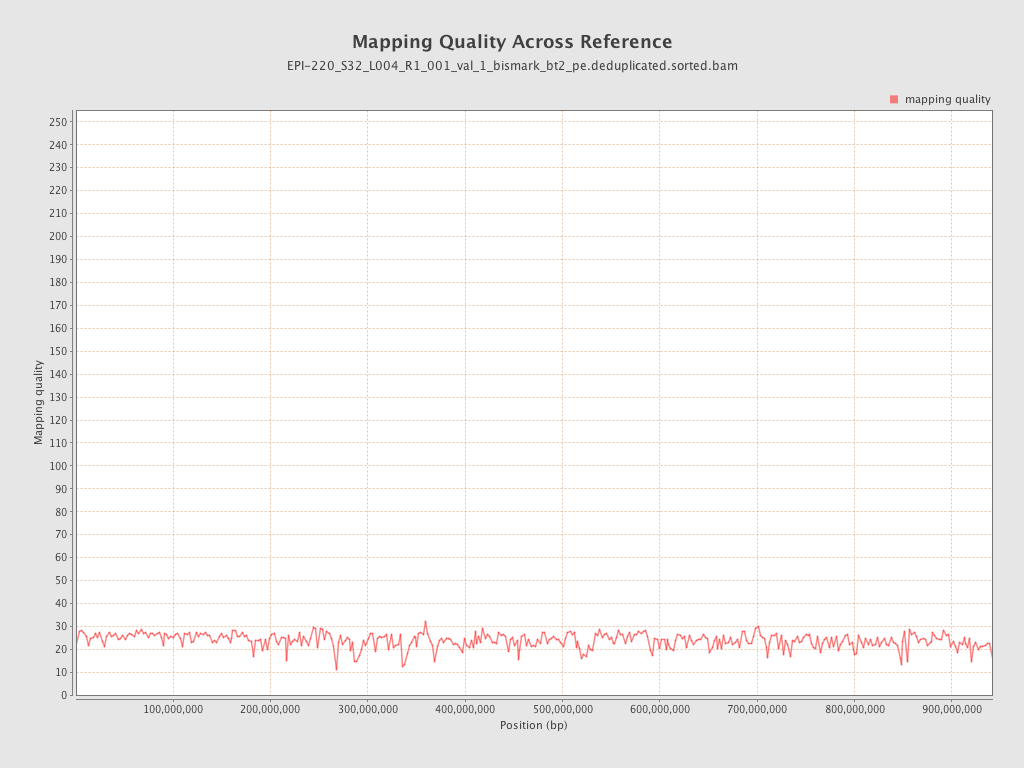
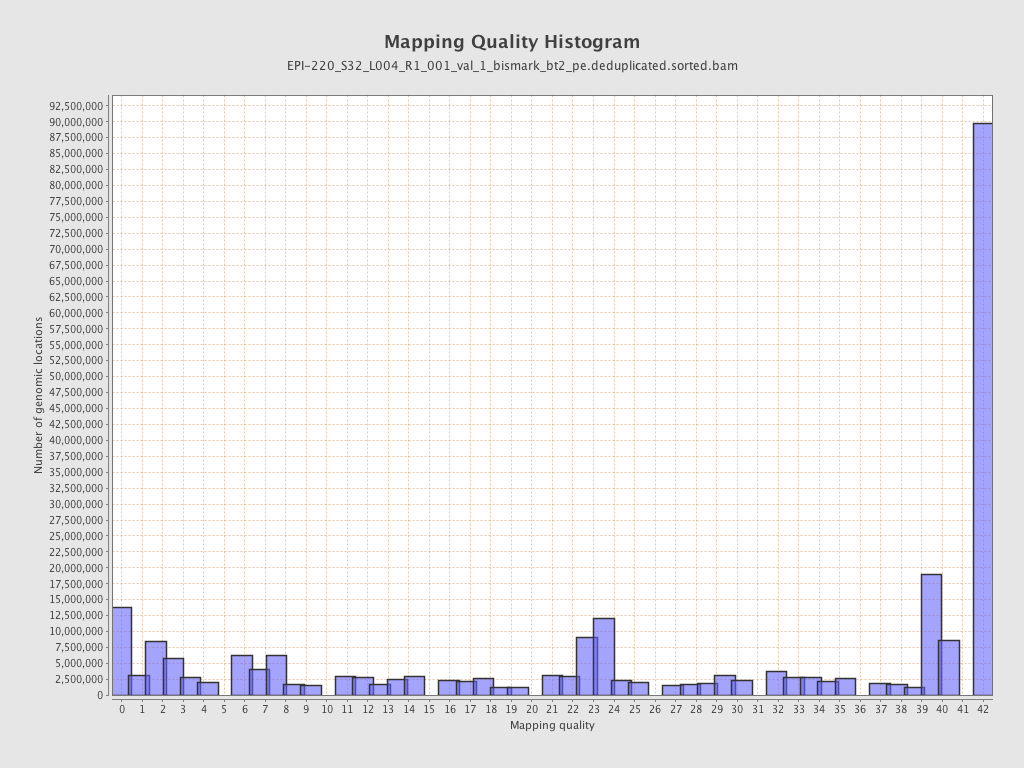
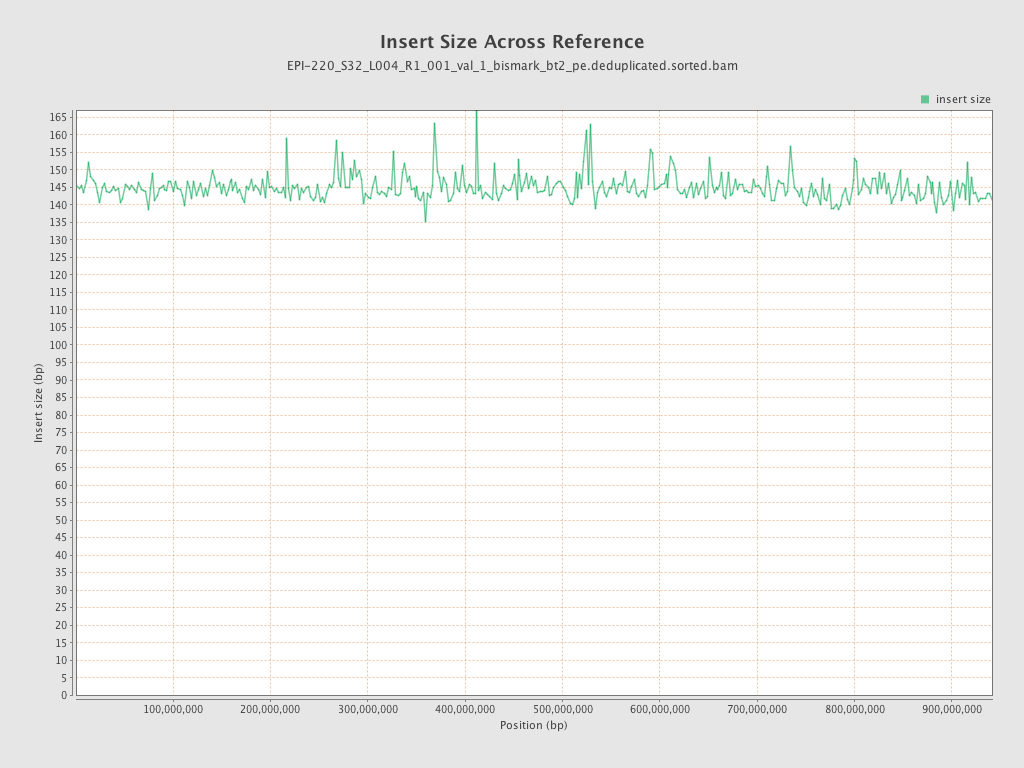
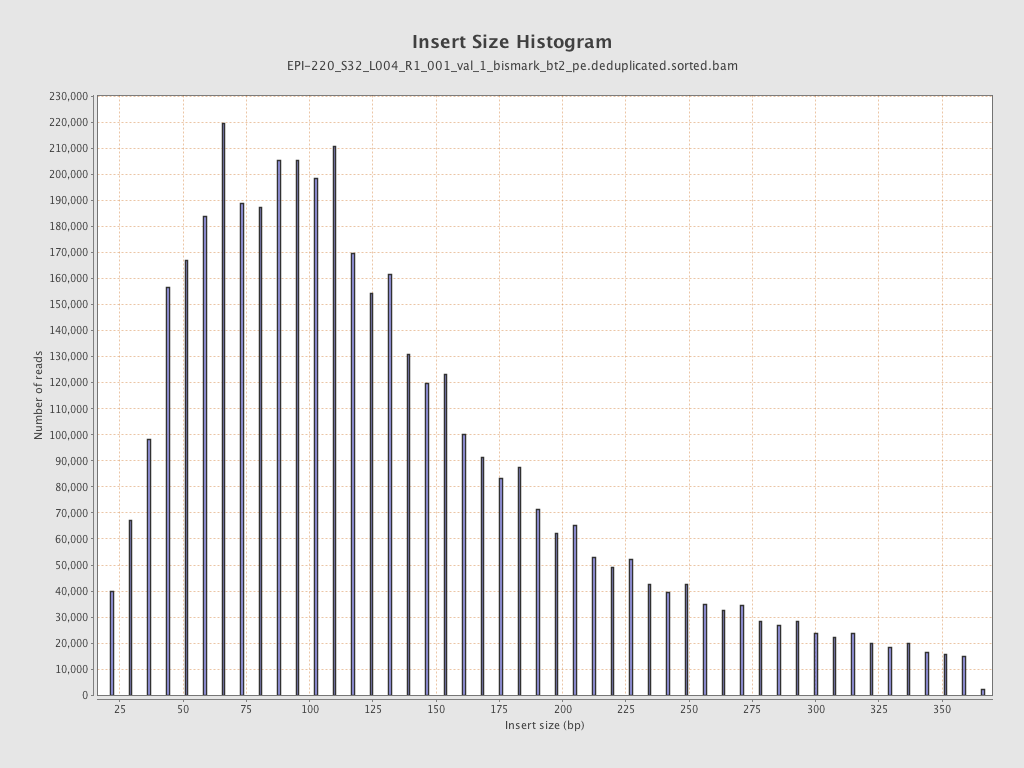
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
76418536 |
0.8525 |
3.0101 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
61054930 |
0.8773 |
4.1254 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
48464744 |
0.8393 |
3.0655 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
51833052 |
0.7939 |
4.6344 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
47185233 |
0.7017 |
3.2175 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
47082271 |
0.7623 |
5.6795 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
33932384 |
0.7869 |
2.9074 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
49198685 |
0.8045 |
3.2467 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
35066239 |
0.9089 |
8.678 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
40562905 |
0.7517 |
2.5393 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
36125731 |
0.7022 |
3.464 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
43487937 |
0.8622 |
4.2434 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
35458918 |
0.7987 |
3.4009 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
47249997 |
1.0409 |
18.2066 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
31385539 |
0.6547 |
2.8765 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
27940426 |
0.8737 |
11.7821 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
28289819 |
0.8101 |
3.4919 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
19982450 |
0.7204 |
4.4754 |