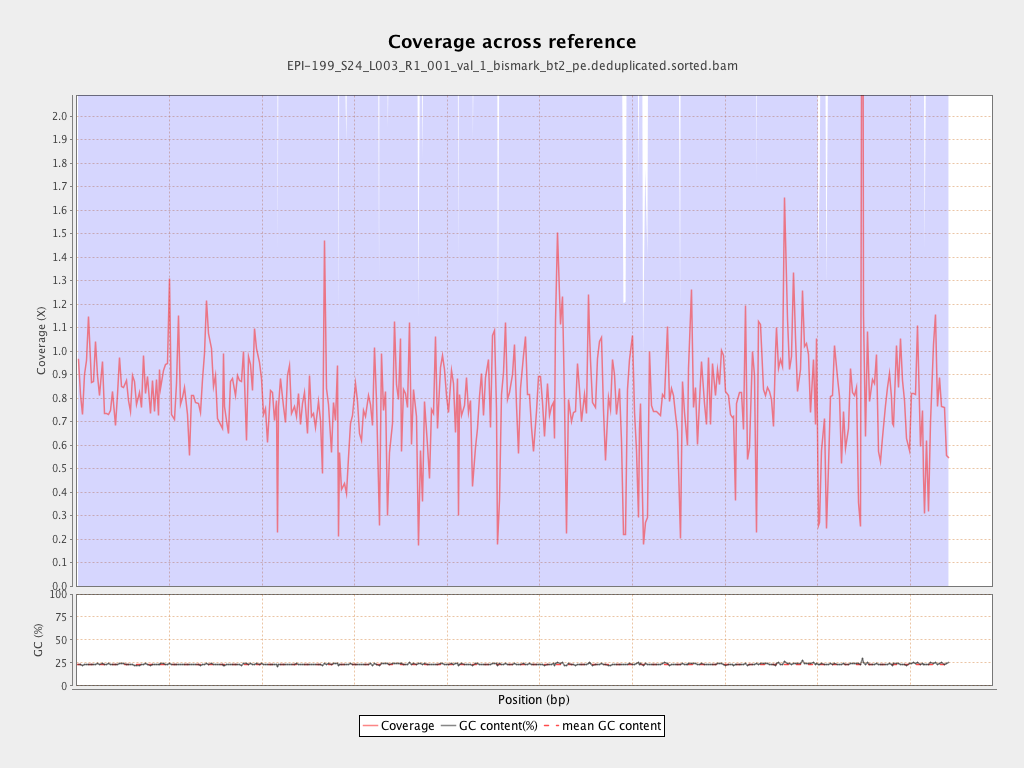
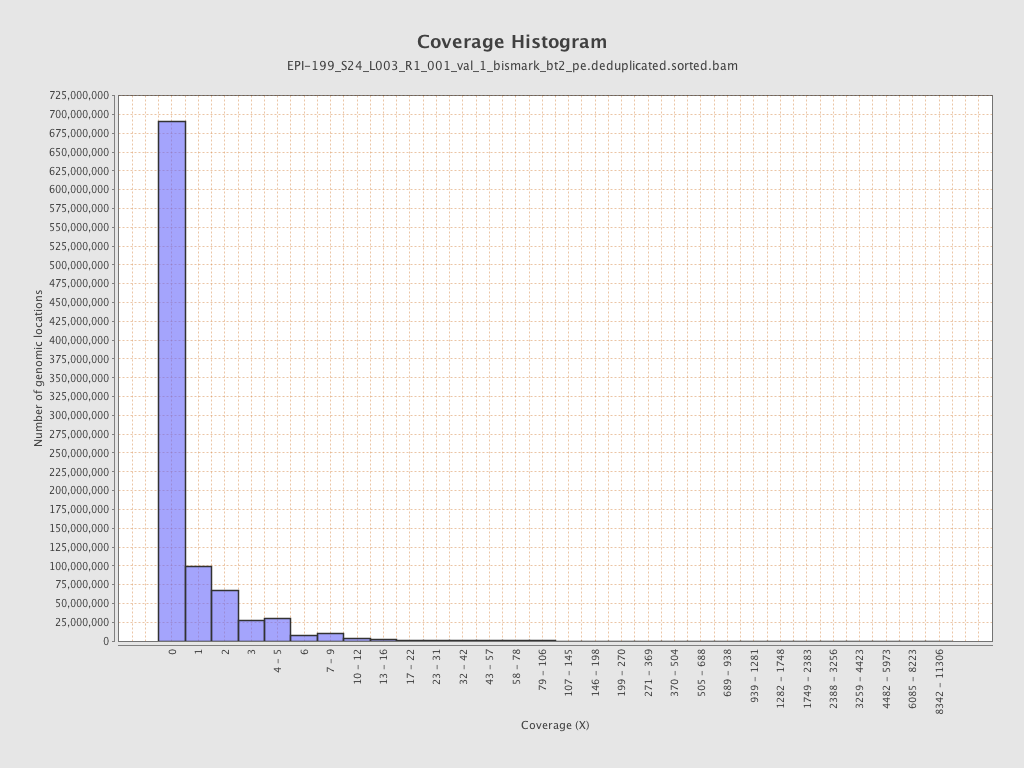
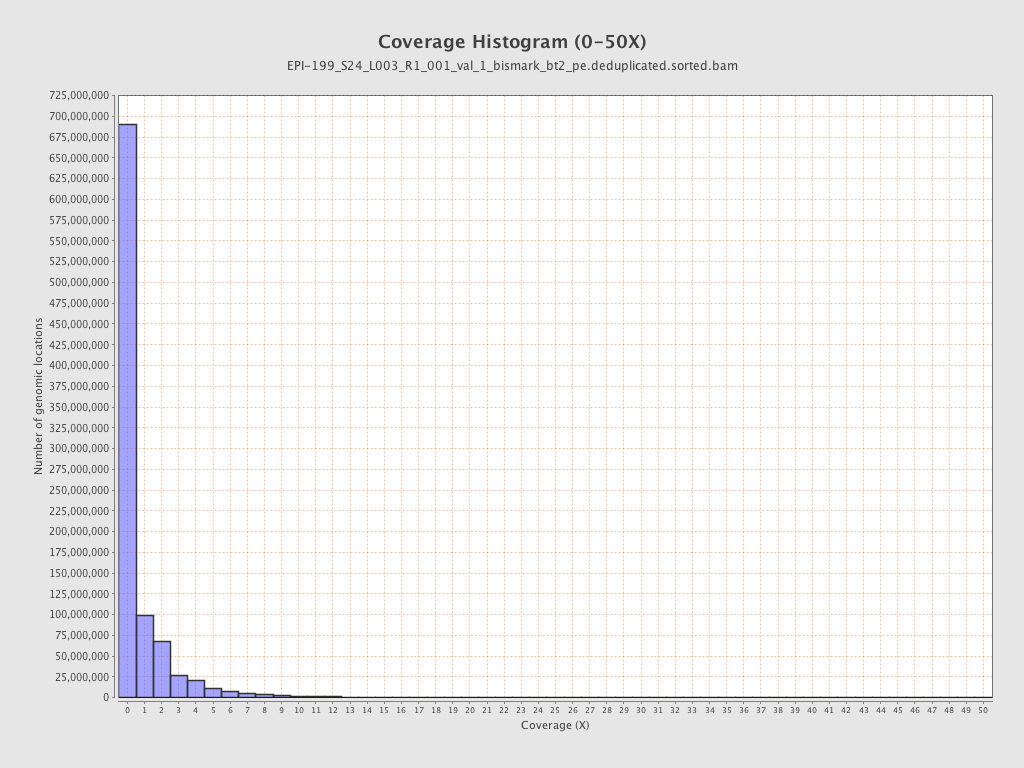
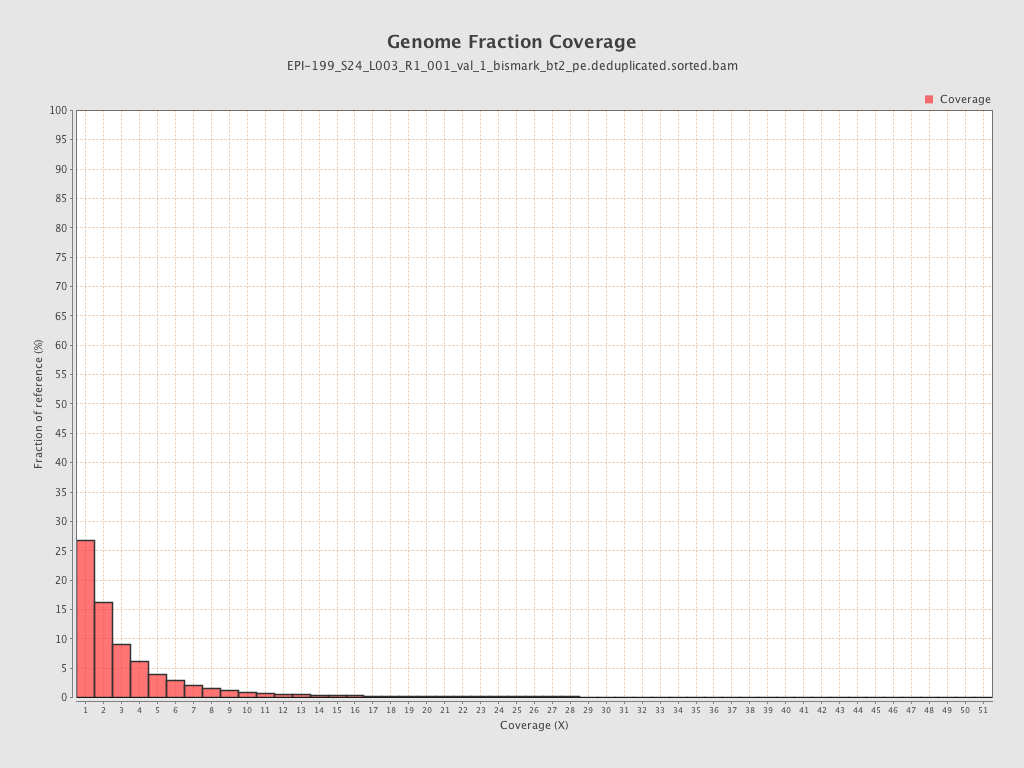
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
75651478 |
0.8439 |
3.0297 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
60244764 |
0.8656 |
4.2721 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
48178627 |
0.8344 |
3.0282 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
51195912 |
0.7842 |
4.2995 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
46635649 |
0.6935 |
3.2095 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
46052049 |
0.7457 |
5.0468 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
33695810 |
0.7814 |
2.7697 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
49036164 |
0.8019 |
3.055 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
34317943 |
0.8895 |
8.0447 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
40614623 |
0.7527 |
2.6174 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
35599159 |
0.6919 |
3.3914 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
42777756 |
0.8481 |
4.1451 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
35326923 |
0.7957 |
3.392 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
46422132 |
1.0227 |
17.7783 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
31261282 |
0.6521 |
2.7941 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
27435488 |
0.8579 |
11.5888 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
28021956 |
0.8024 |
3.3519 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
19631530 |
0.7078 |
4.9052 |