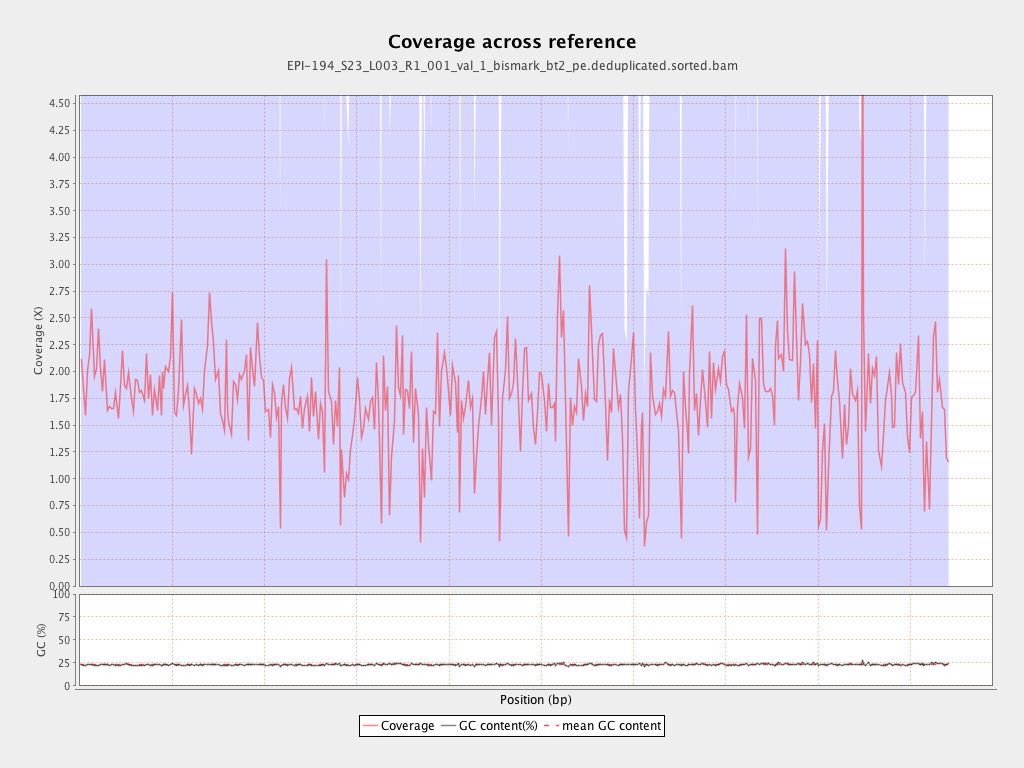
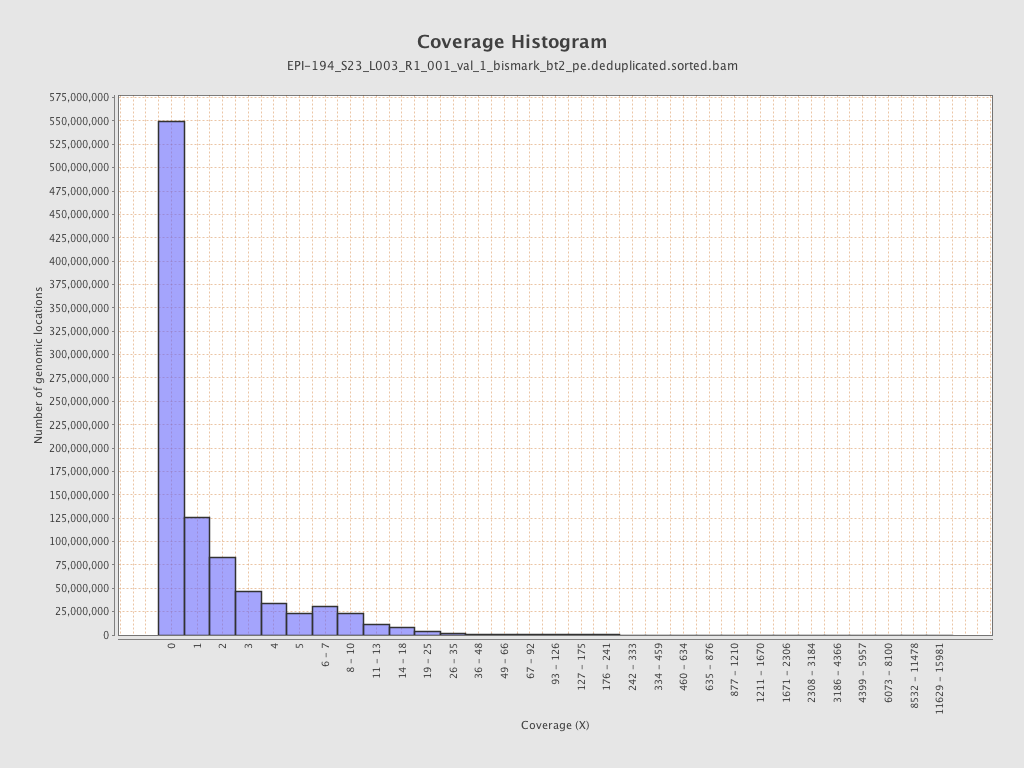
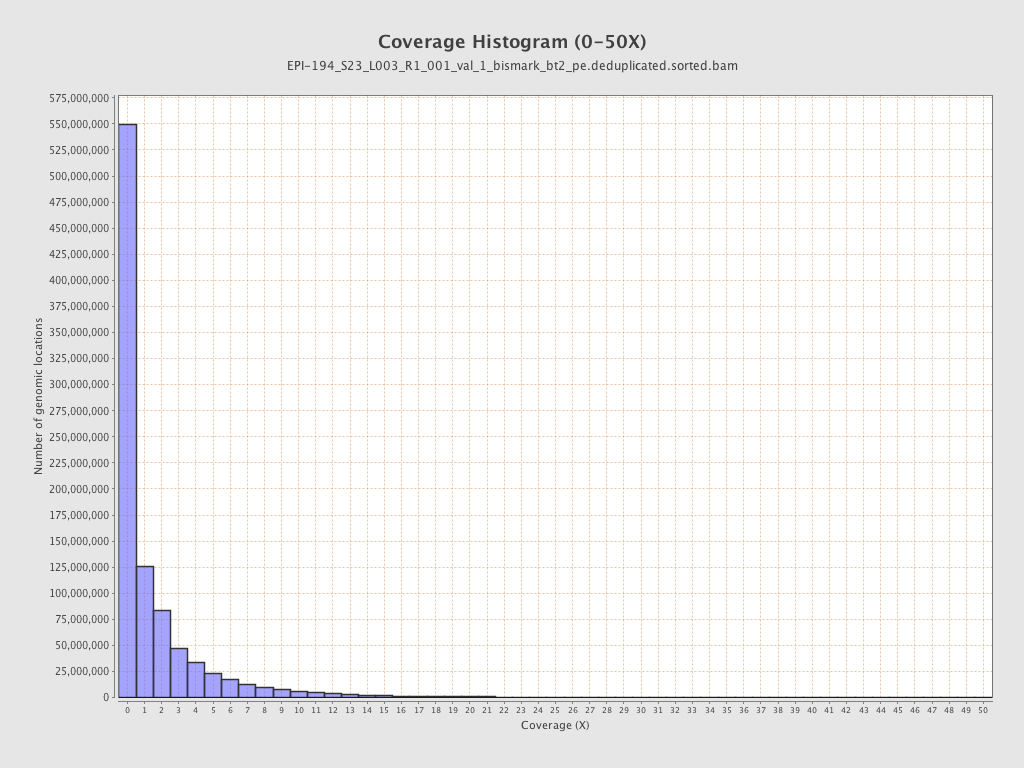
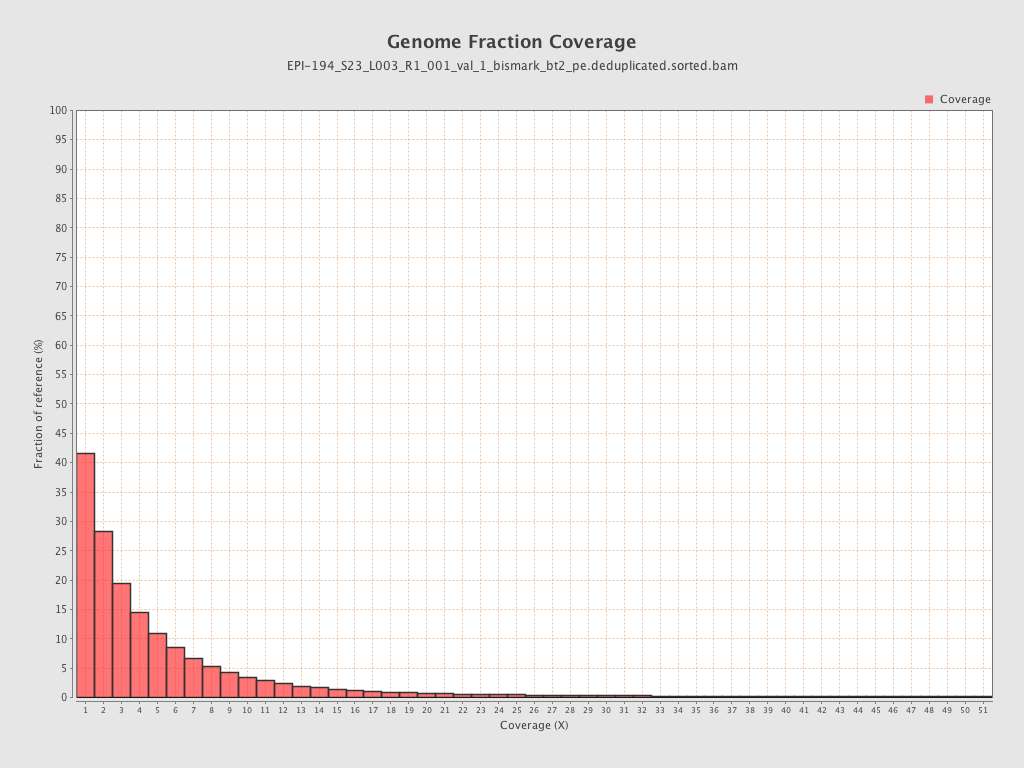
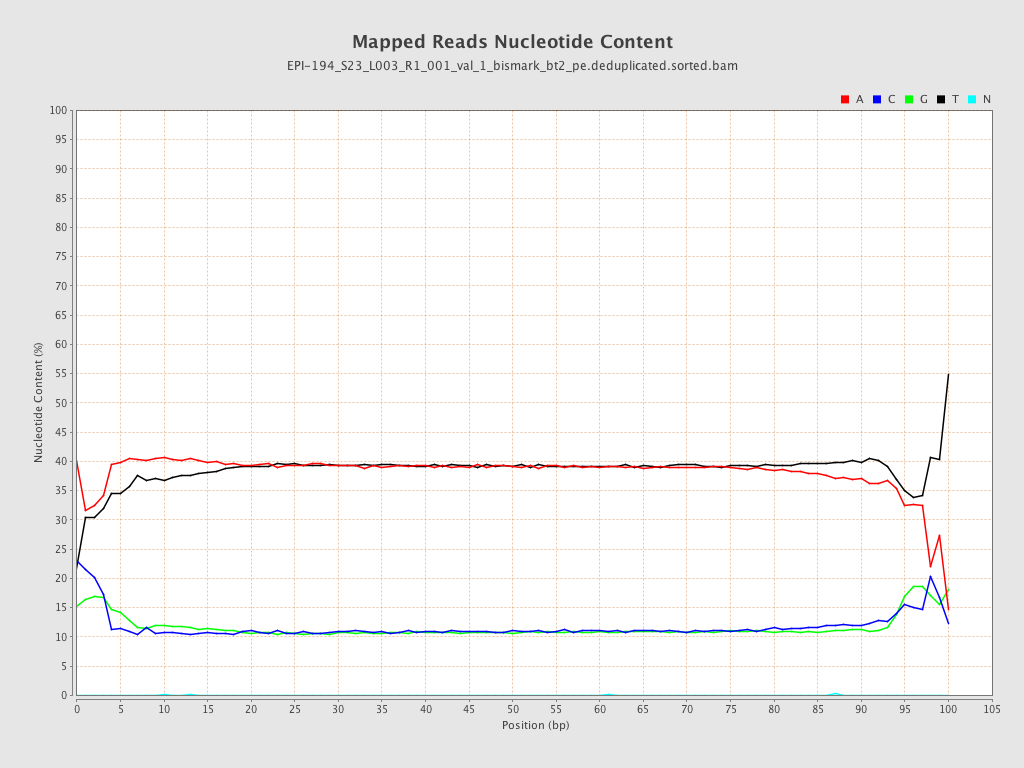
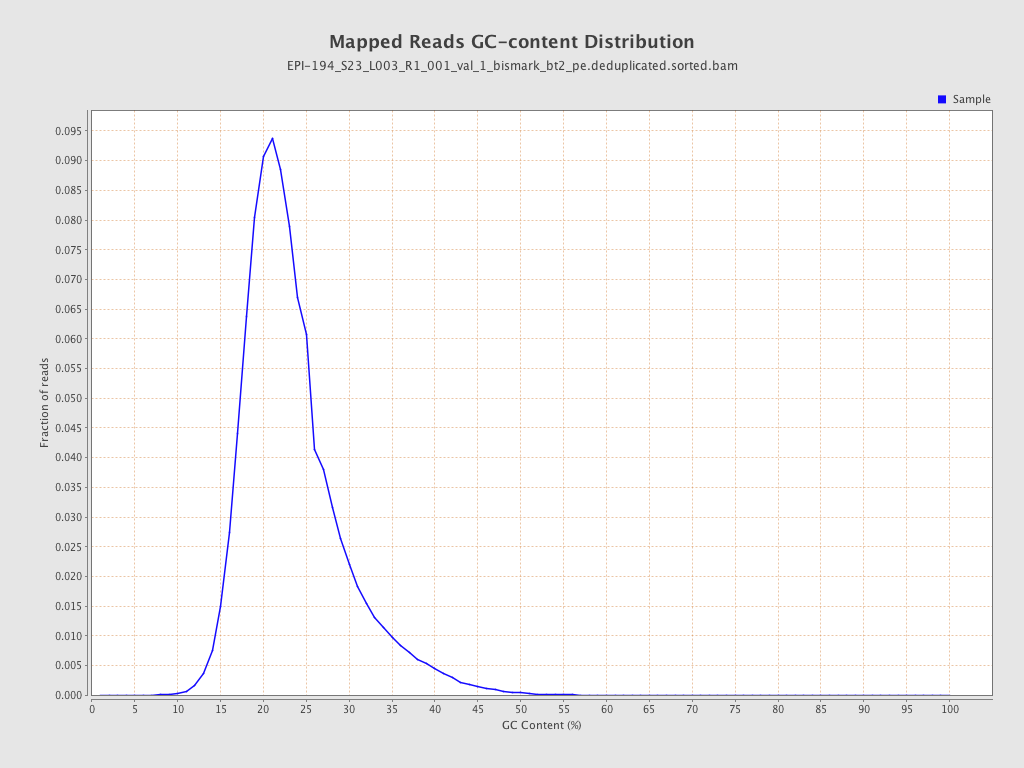
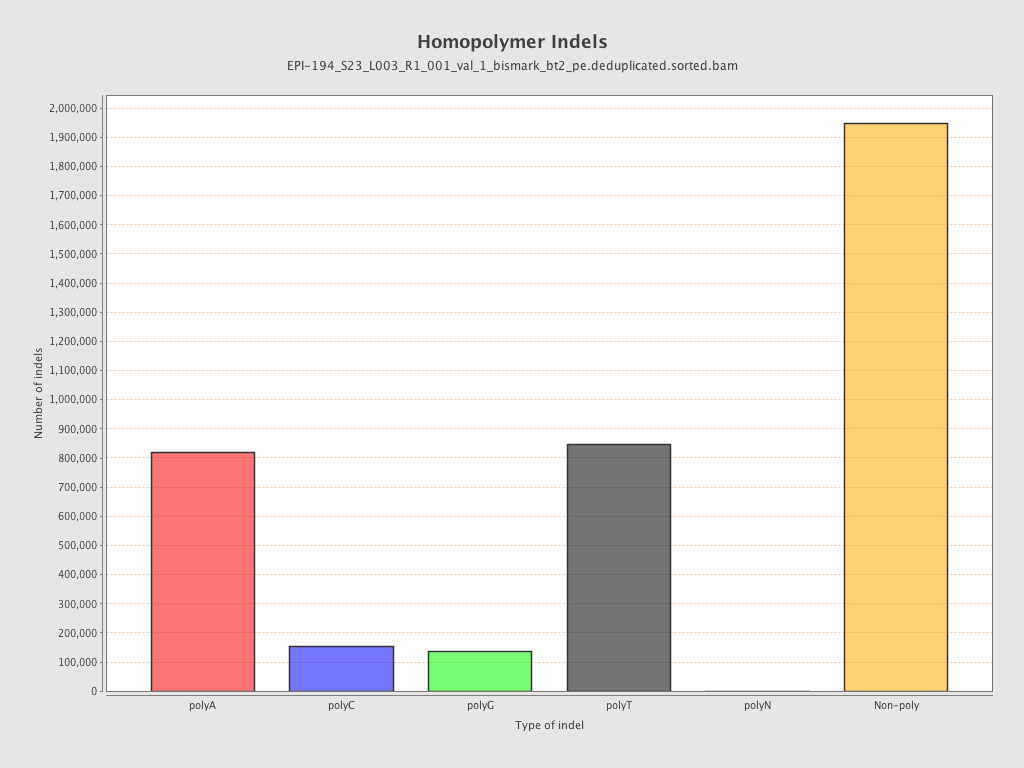
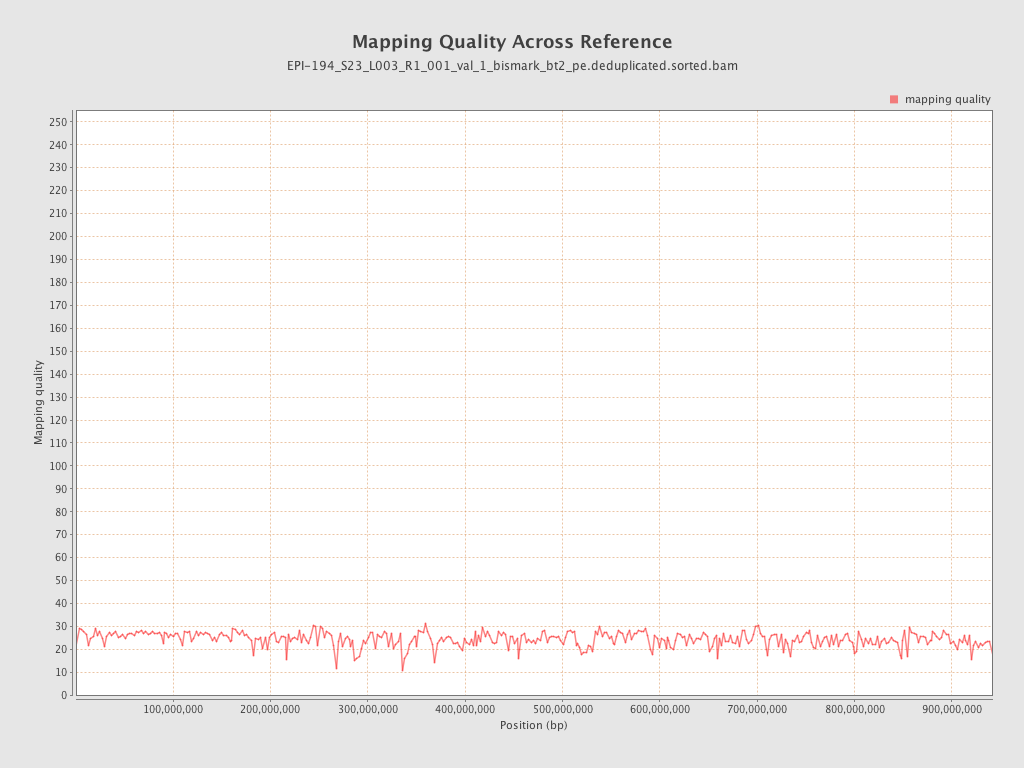
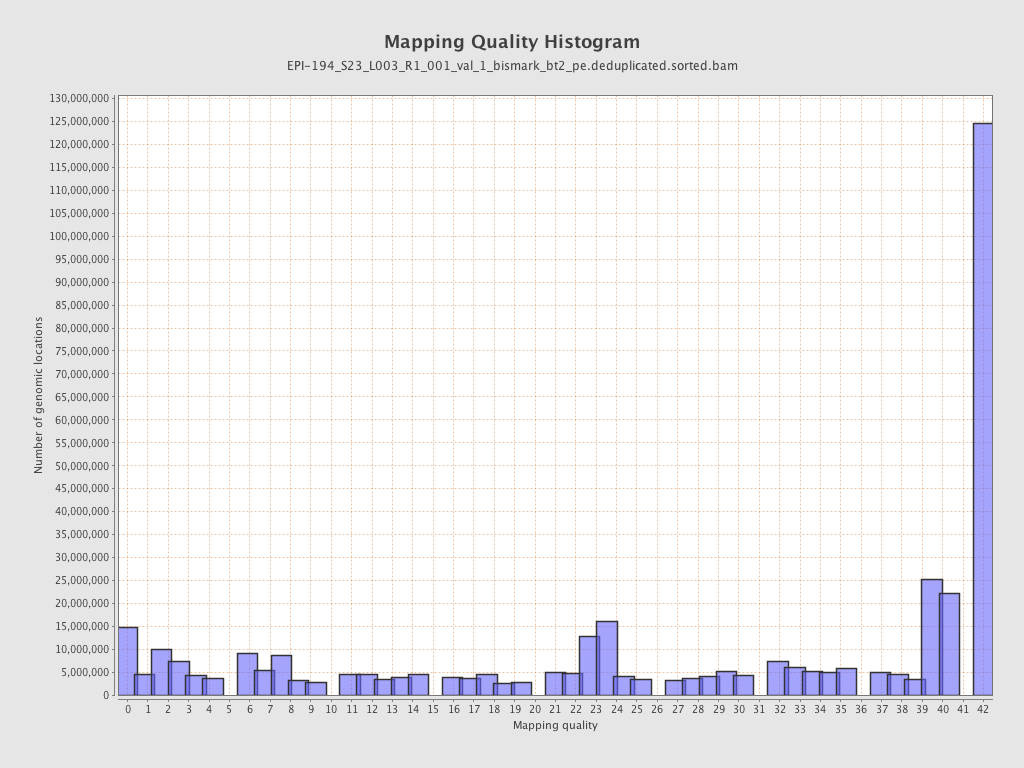
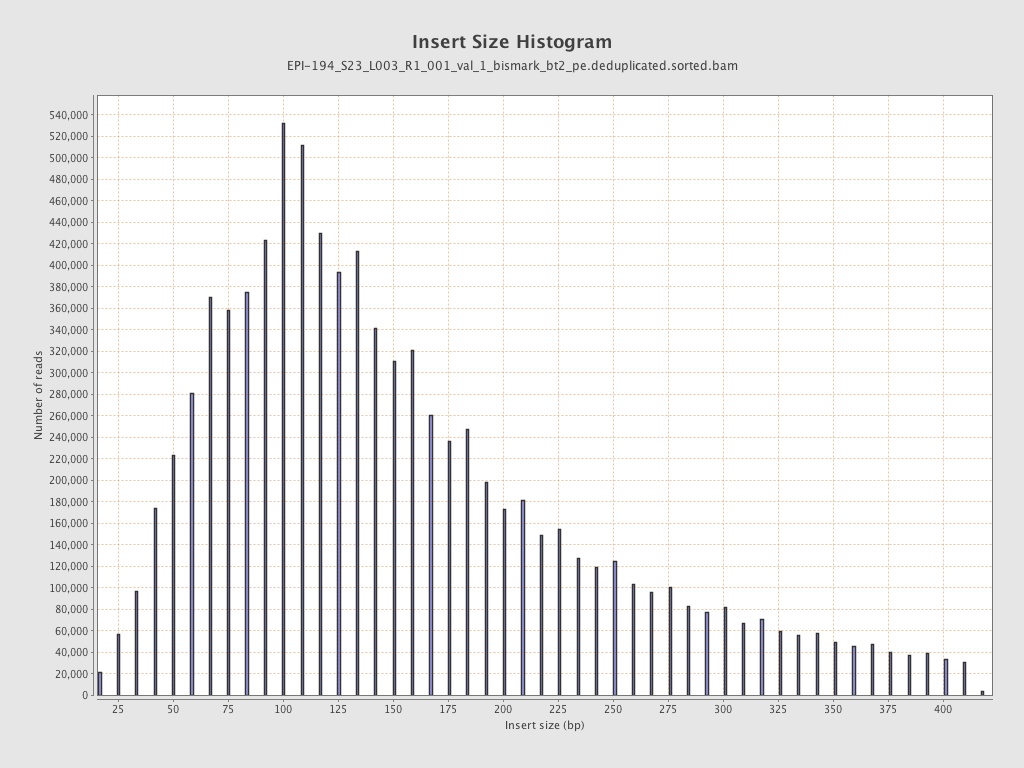
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
168568105 |
1.8804 |
5.3619 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
133687819 |
1.9209 |
7.3441 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
105568776 |
1.8282 |
5.792 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
111887540 |
1.7137 |
8.1465 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
102442478 |
1.5233 |
5.7034 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
101075519 |
1.6366 |
7.9753 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
73408585 |
1.7024 |
5.2997 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
107817735 |
1.7631 |
5.939 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
74885949 |
1.941 |
14.2886 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
90309822 |
1.6736 |
4.8988 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
78759230 |
1.5308 |
6.4614 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
94089988 |
1.8654 |
7.3688 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
77509910 |
1.7458 |
6.5551 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
100656219 |
2.2174 |
26.0585 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
68217120 |
1.423 |
5.2942 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
59117990 |
1.8485 |
16.2646 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
61178097 |
1.7518 |
5.8598 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
43119263 |
1.5545 |
9.2436 |