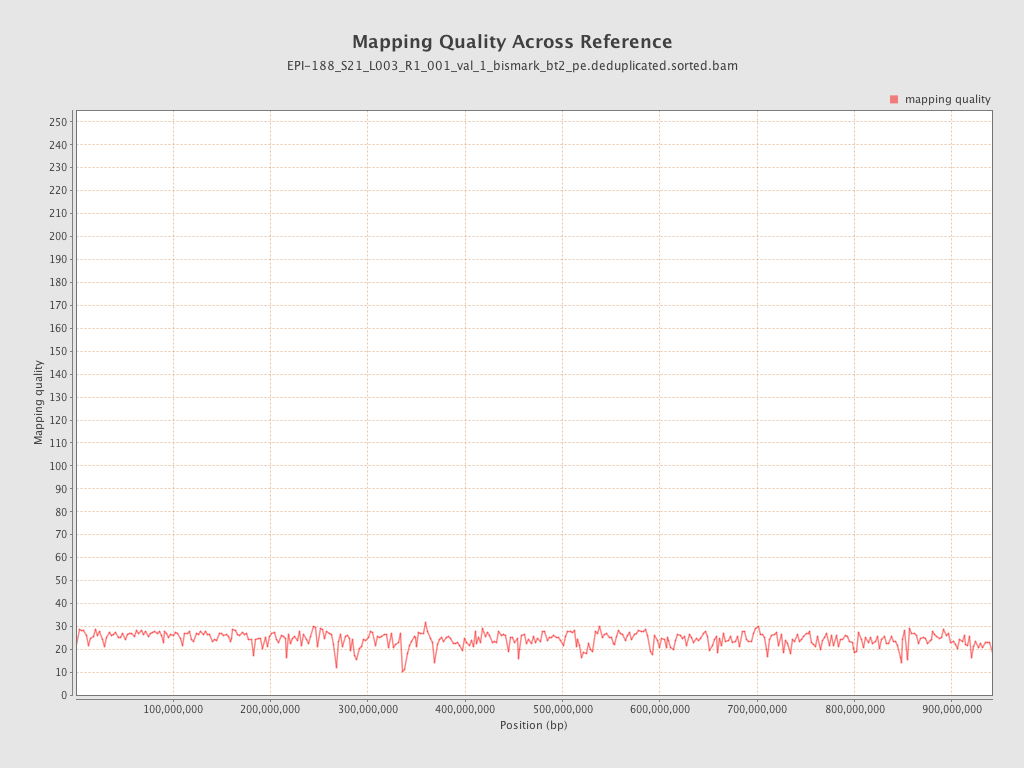
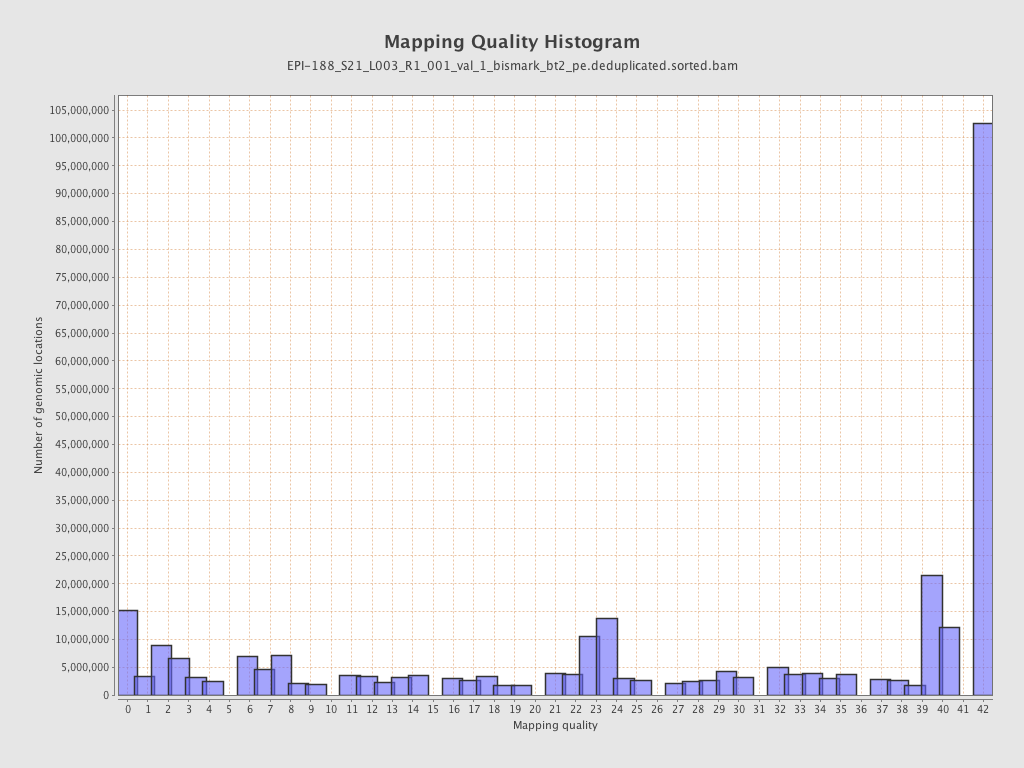
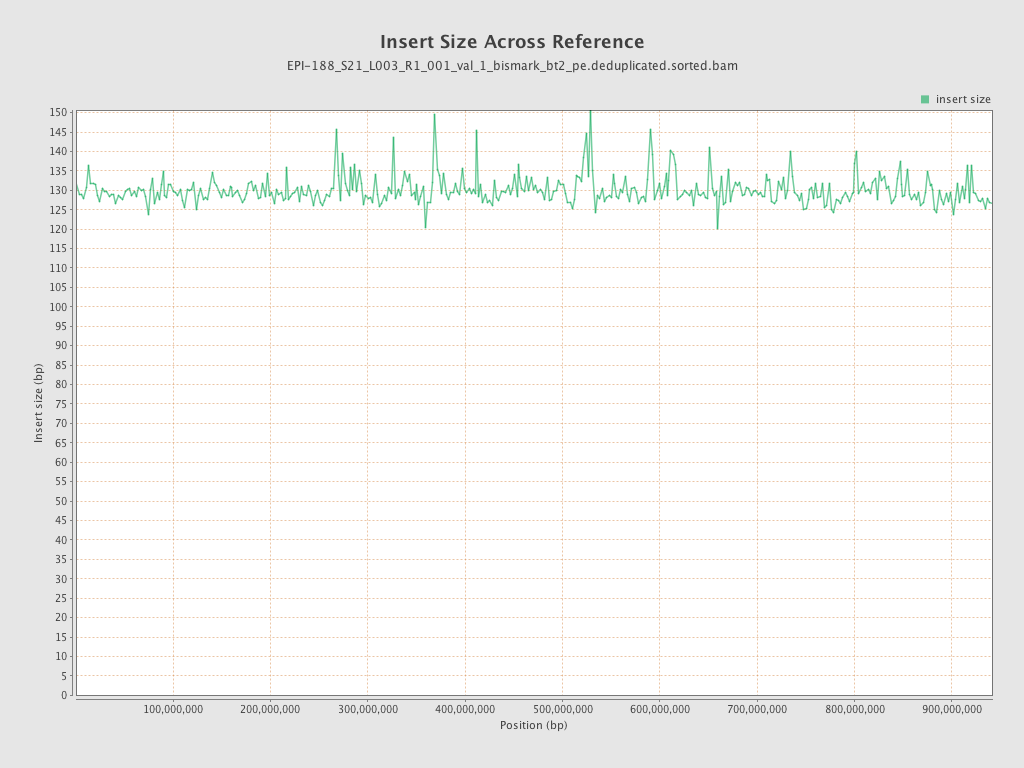
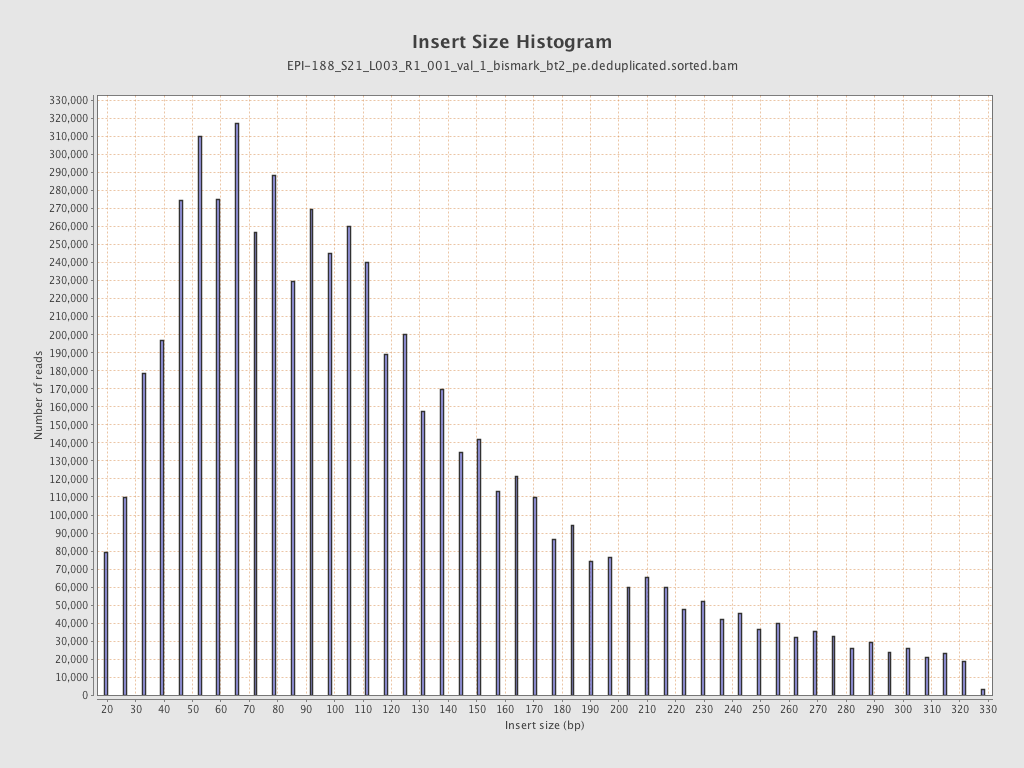
| Name |
Length |
Mapped bases |
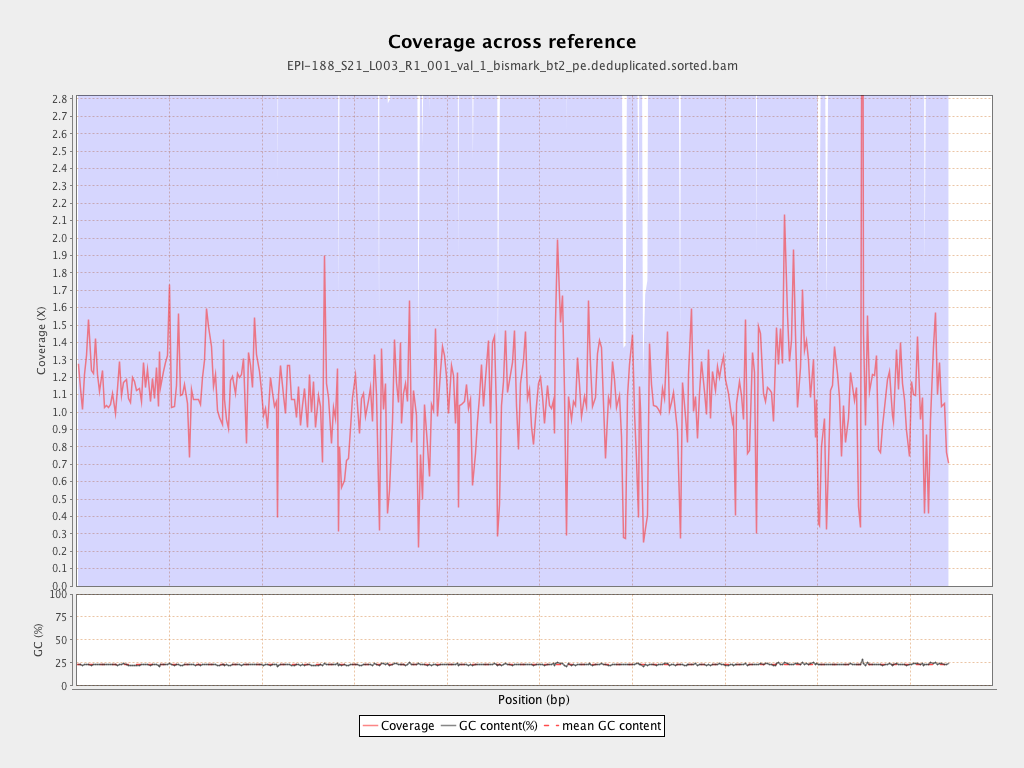
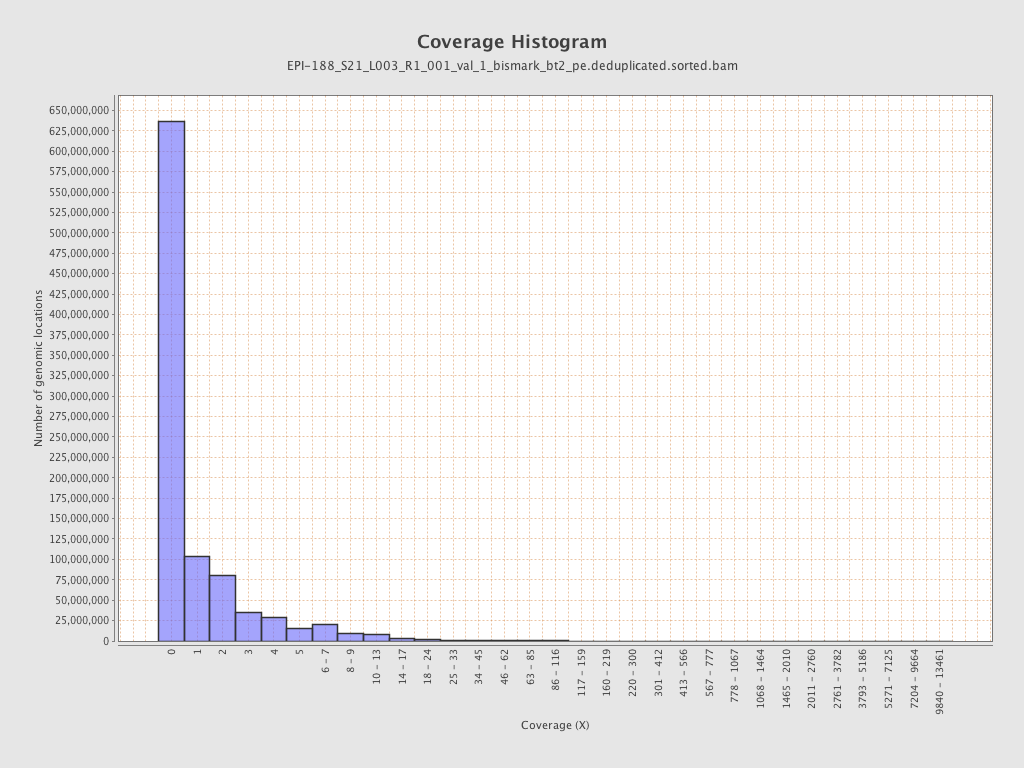
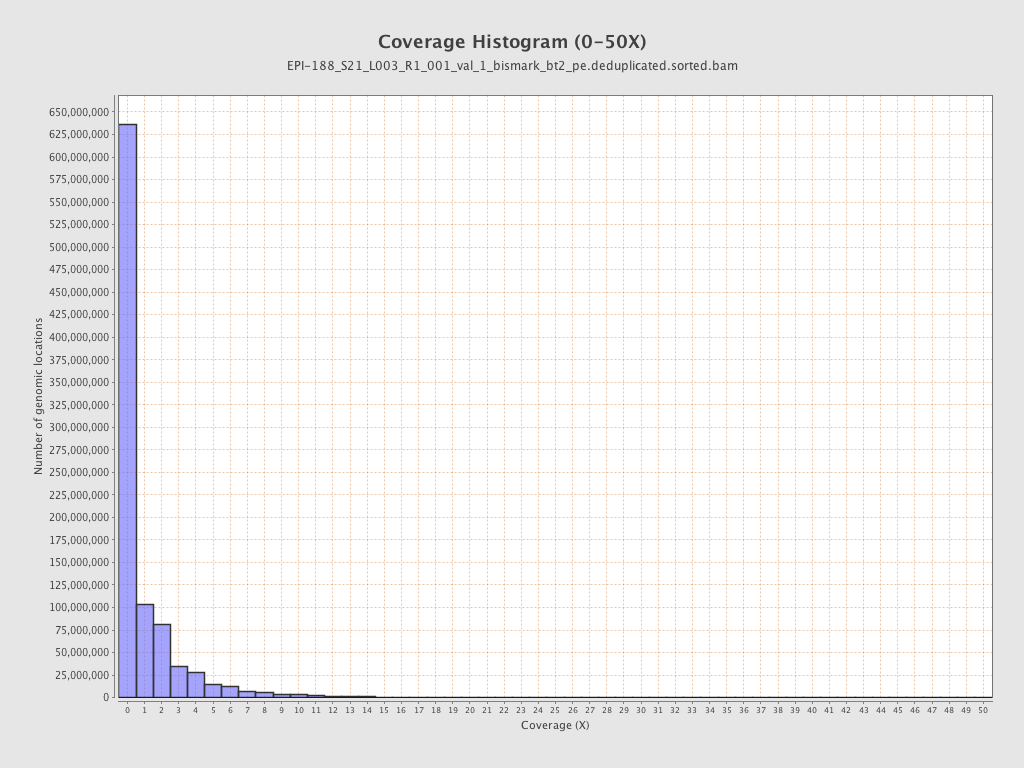
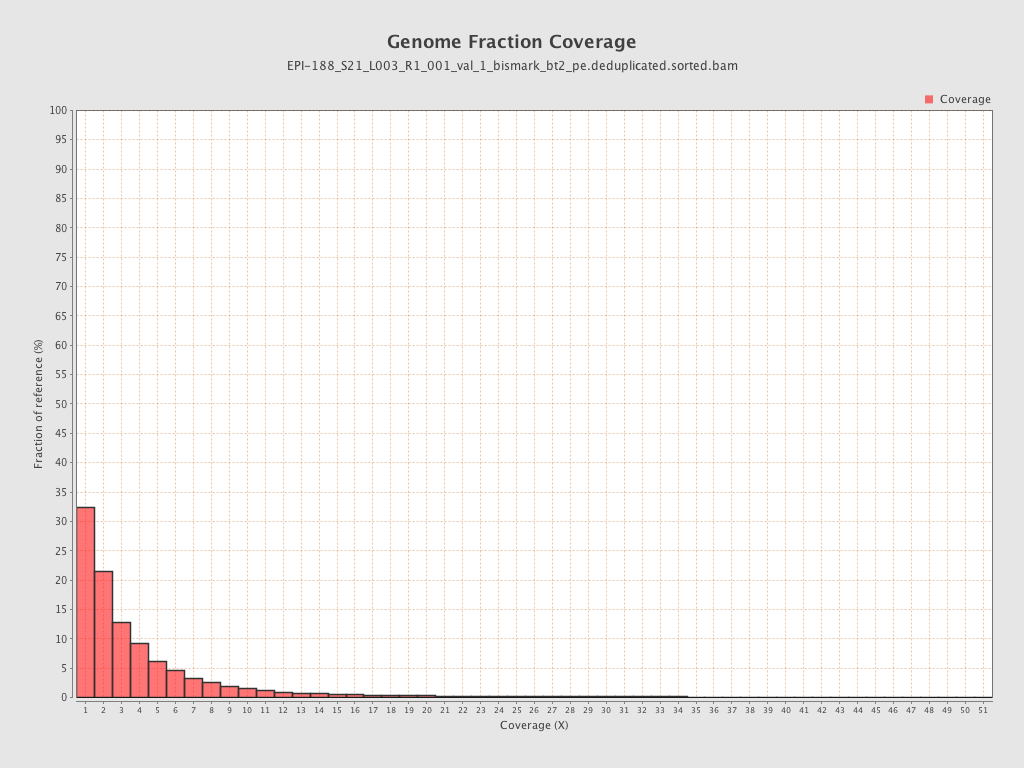
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
104243013 |
1.1629 |
3.8957 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
82564157 |
1.1863 |
5.342 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
65696196 |
1.1377 |
3.6593 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
71244067 |
1.0912 |
5.6387 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
64231405 |
0.9551 |
3.9737 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
64103338 |
1.0379 |
7.1382 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
45996307 |
1.0667 |
3.4606 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
67015097 |
1.0959 |
3.9912 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
46920811 |
1.2161 |
10.2774 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
54672630 |
1.0132 |
3.1107 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
48524375 |
0.9431 |
3.941 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
58008705 |
1.1501 |
4.8388 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
47602343 |
1.0722 |
4.1622 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
63070384 |
1.3894 |
20.3313 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
42819577 |
0.8932 |
3.4677 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
37803357 |
1.1821 |
14.3232 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
38391893 |
1.0993 |
4.2905 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
27014826 |
0.9739 |
6.1144 |