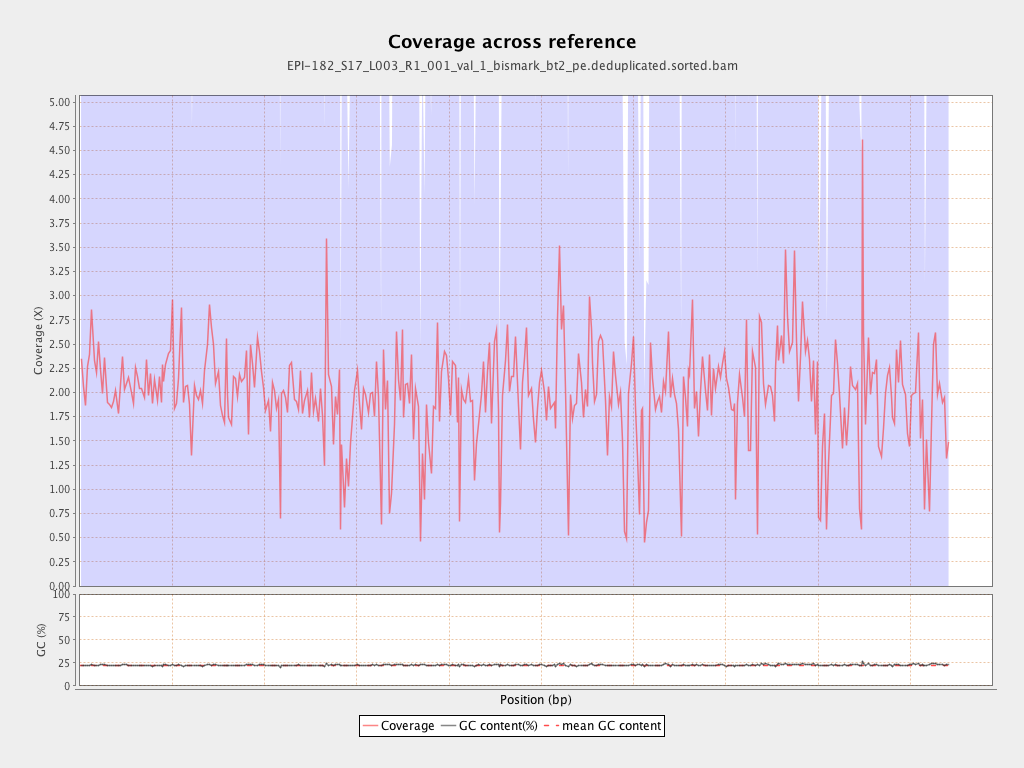
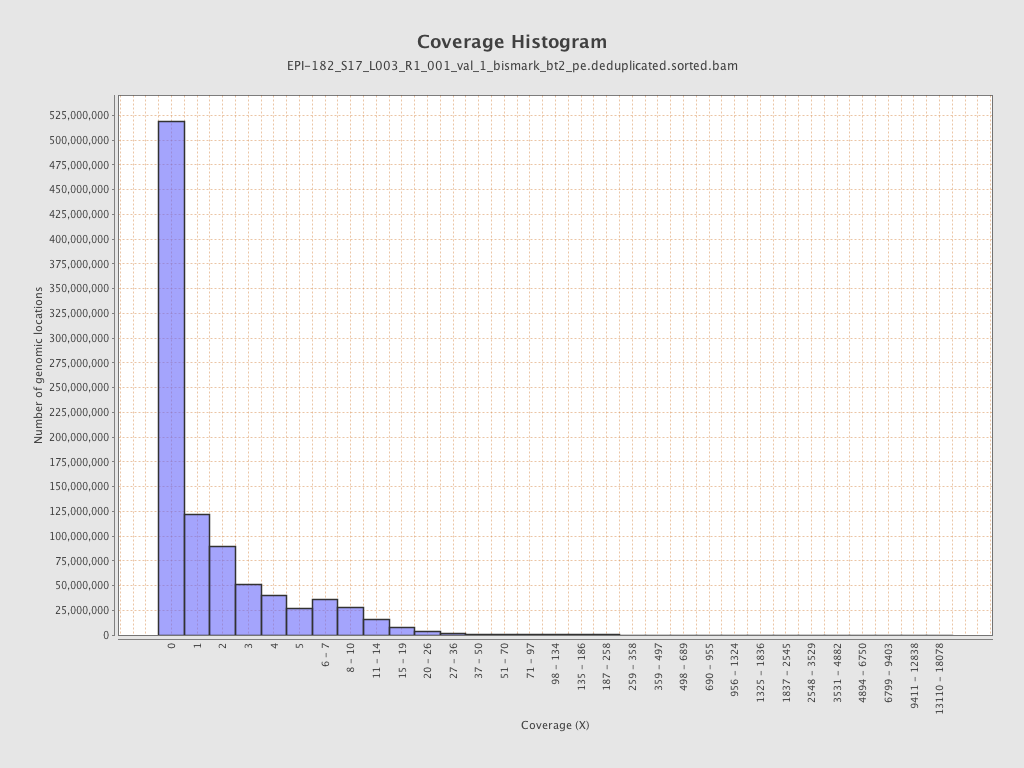
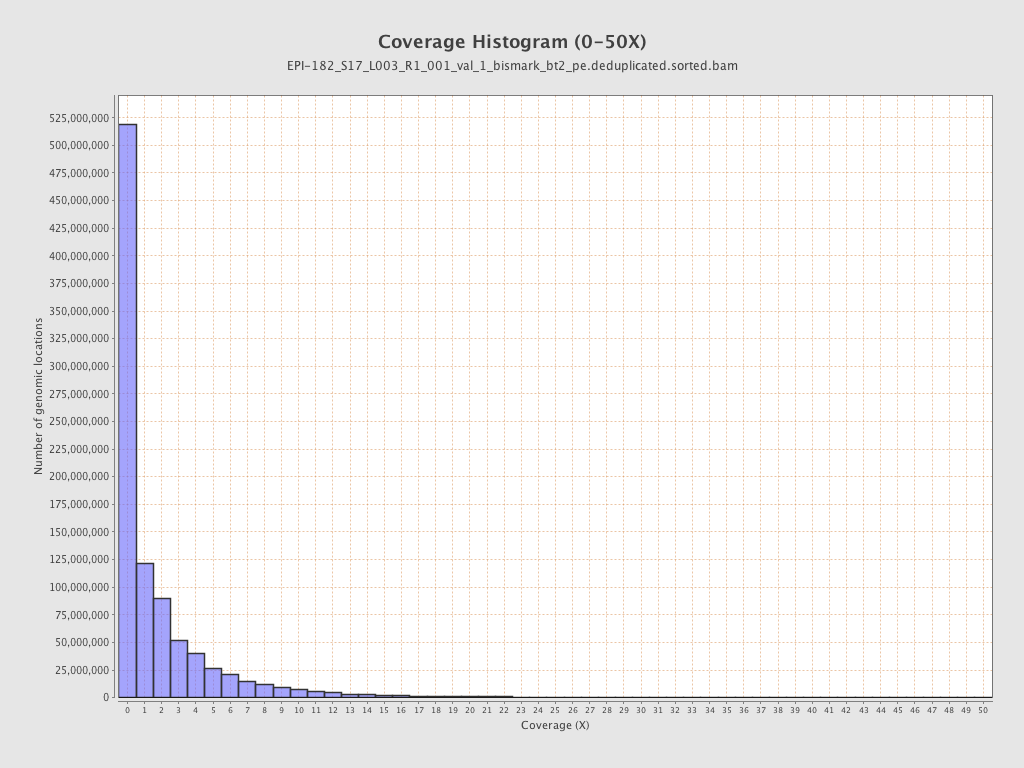
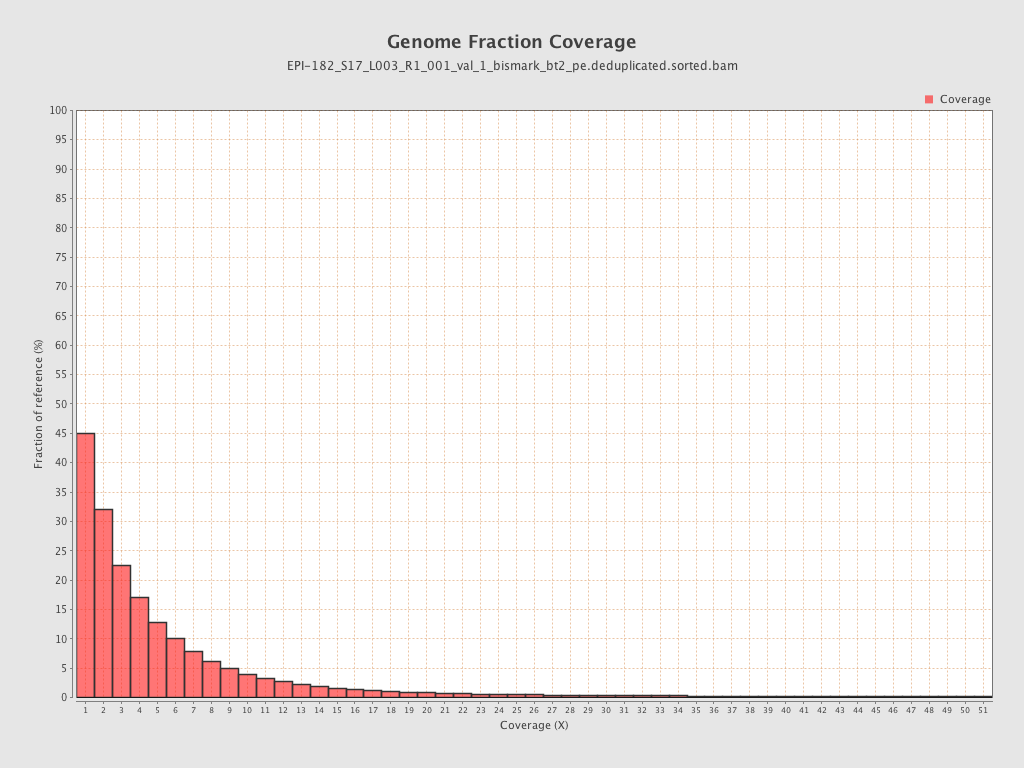
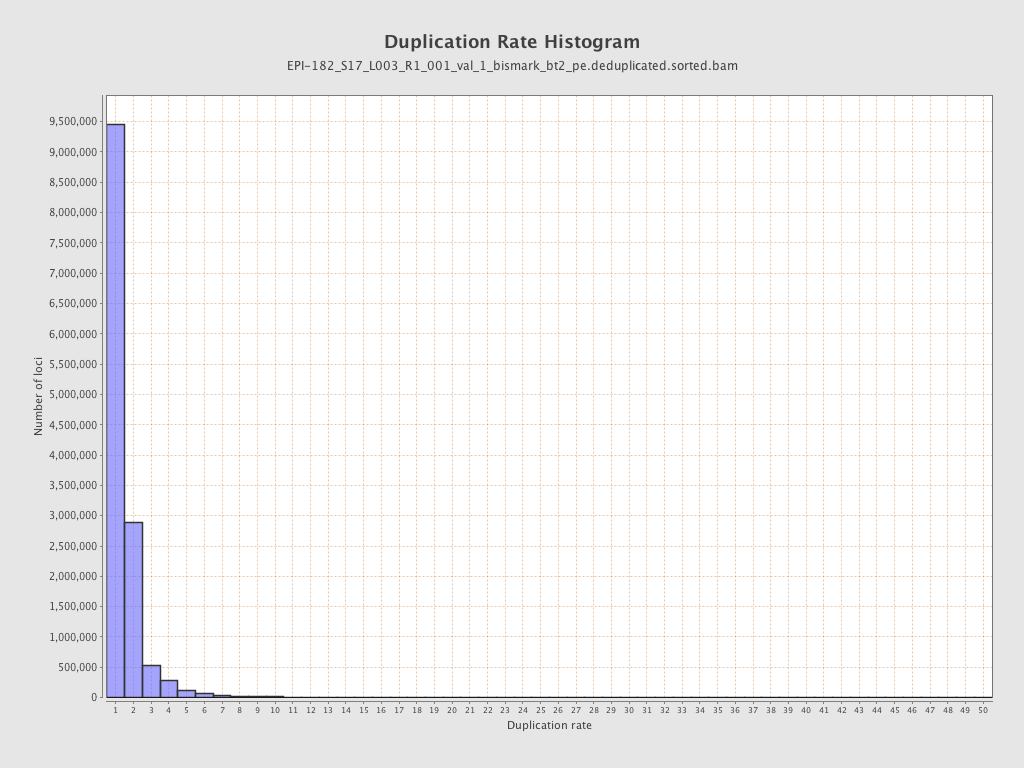
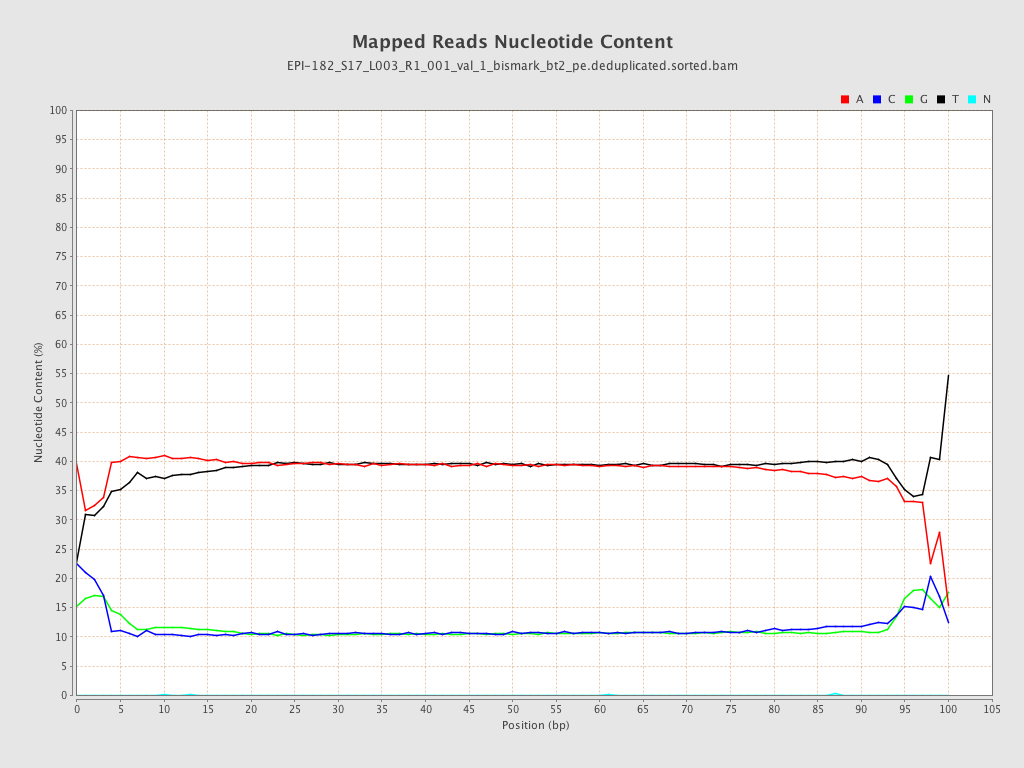
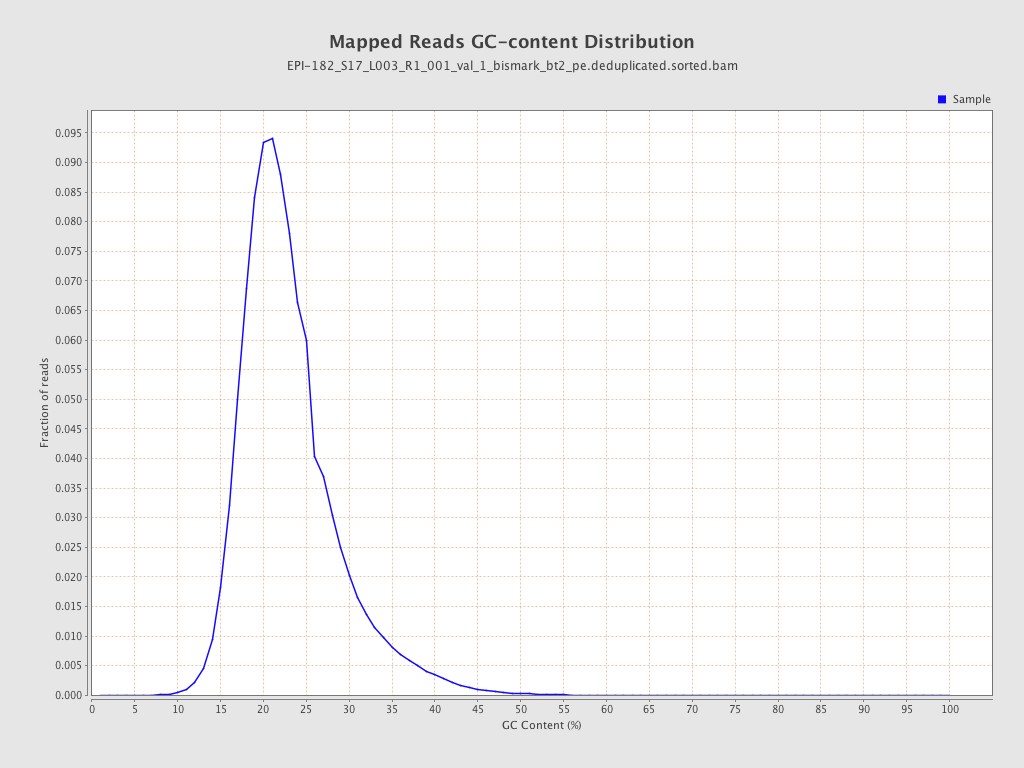
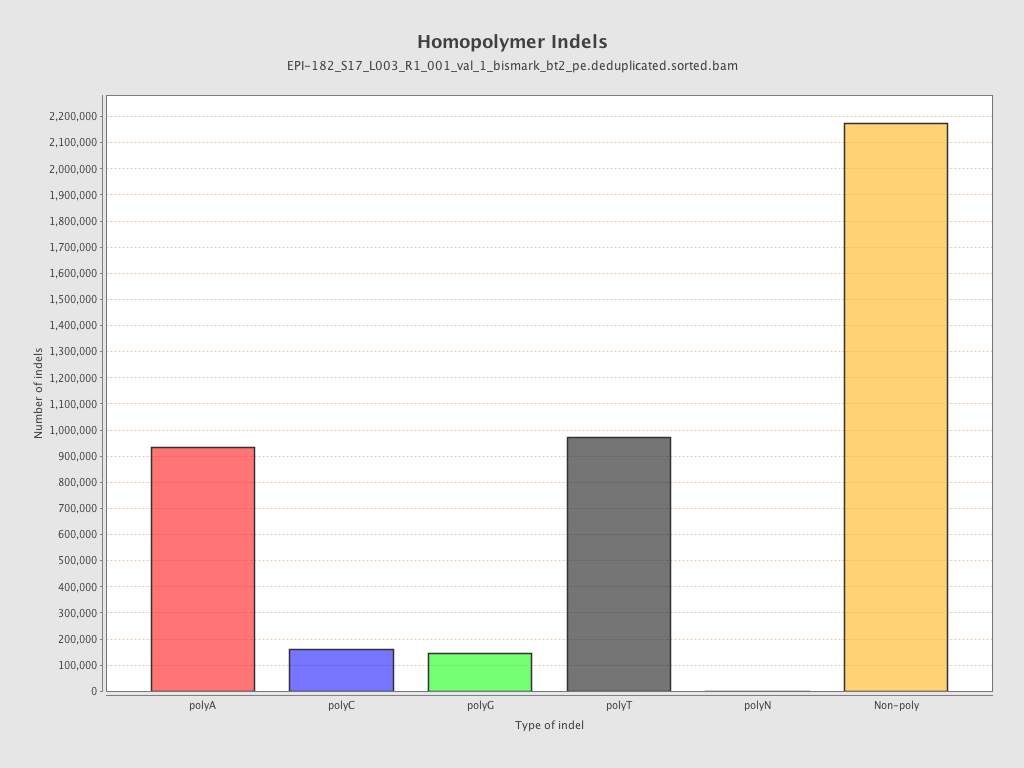
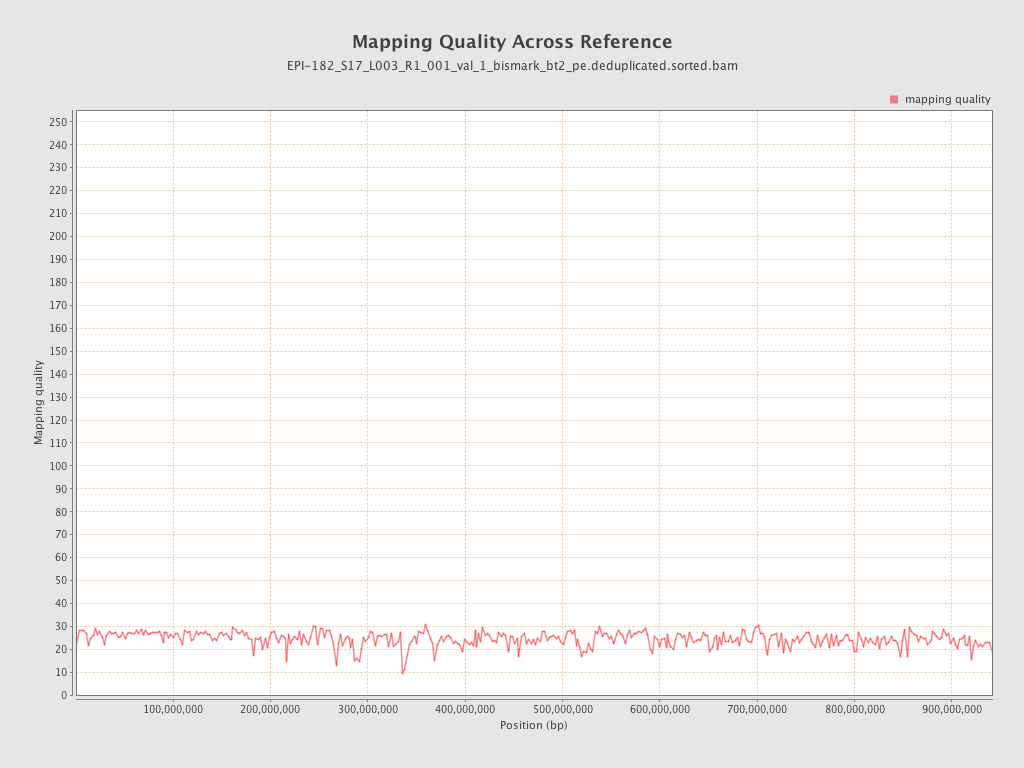
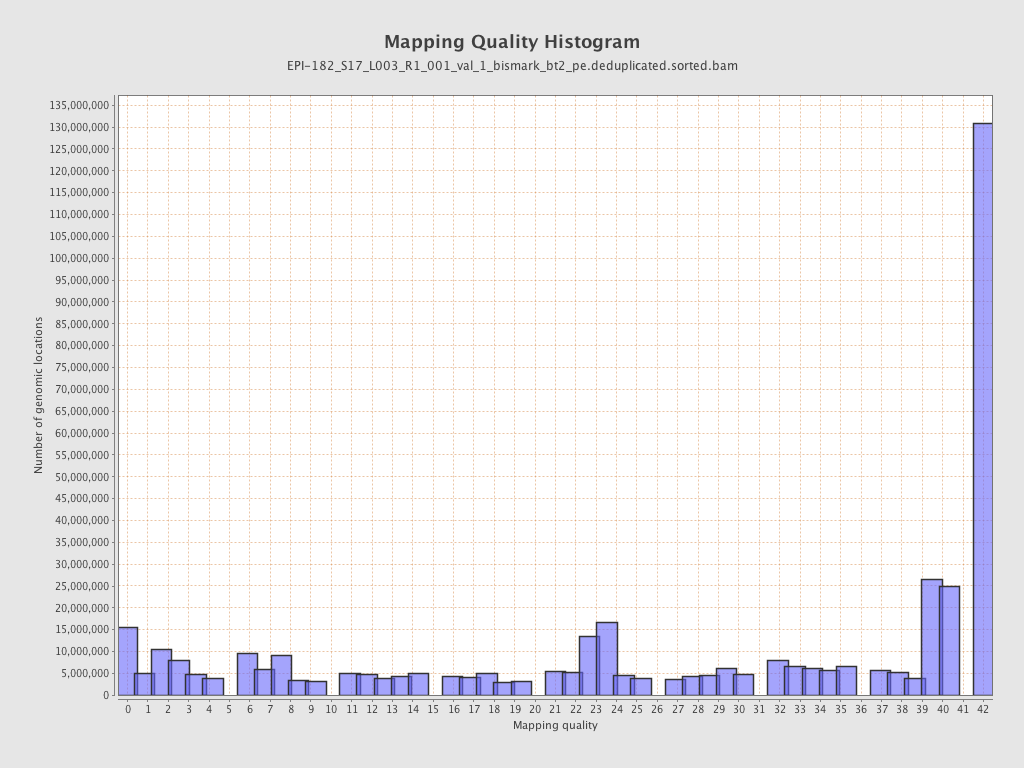
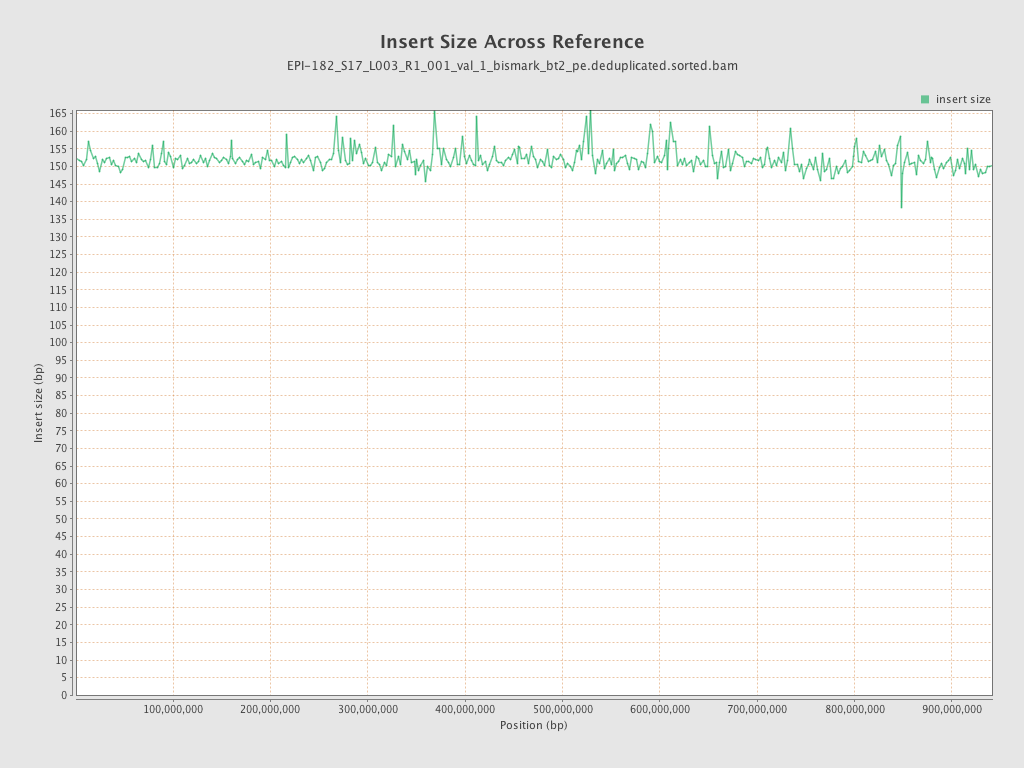
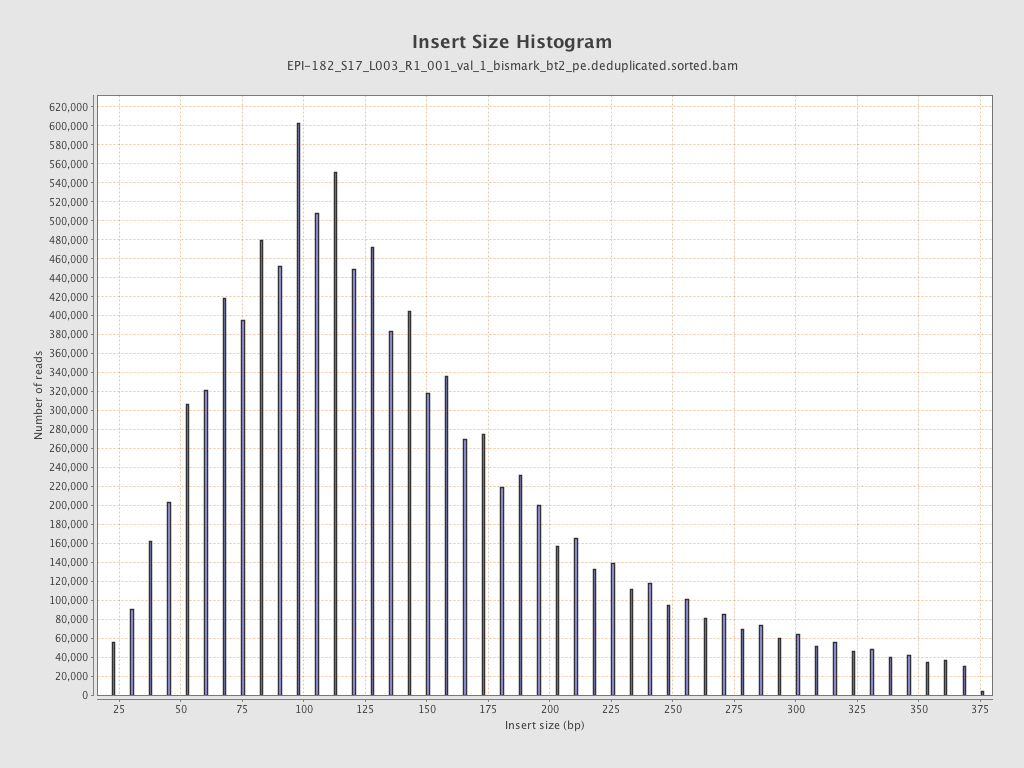
| Name |
Length |
Mapped bases |
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
189546243 |
2.1144 |
5.6458 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
150475428 |
2.1621 |
7.5608 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
118522432 |
2.0526 |
6.0709 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
129643935 |
1.9857 |
9.2621 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
116742679 |
1.736 |
6.3138 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
114927620 |
1.8609 |
8.3348 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
83918716 |
1.9462 |
6.0252 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
121756328 |
1.9911 |
6.6267 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
84308533 |
2.1852 |
16.6239 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
99944181 |
1.8521 |
5.0926 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
89908714 |
1.7475 |
6.5516 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
105897682 |
2.0995 |
8.0871 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
86892569 |
1.9572 |
7.0606 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
112964109 |
2.4886 |
28.4904 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
78067968 |
1.6285 |
5.7574 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
65919513 |
2.0612 |
13.8076 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
68818395 |
1.9705 |
5.7829 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
48050100 |
1.7323 |
8.316 |