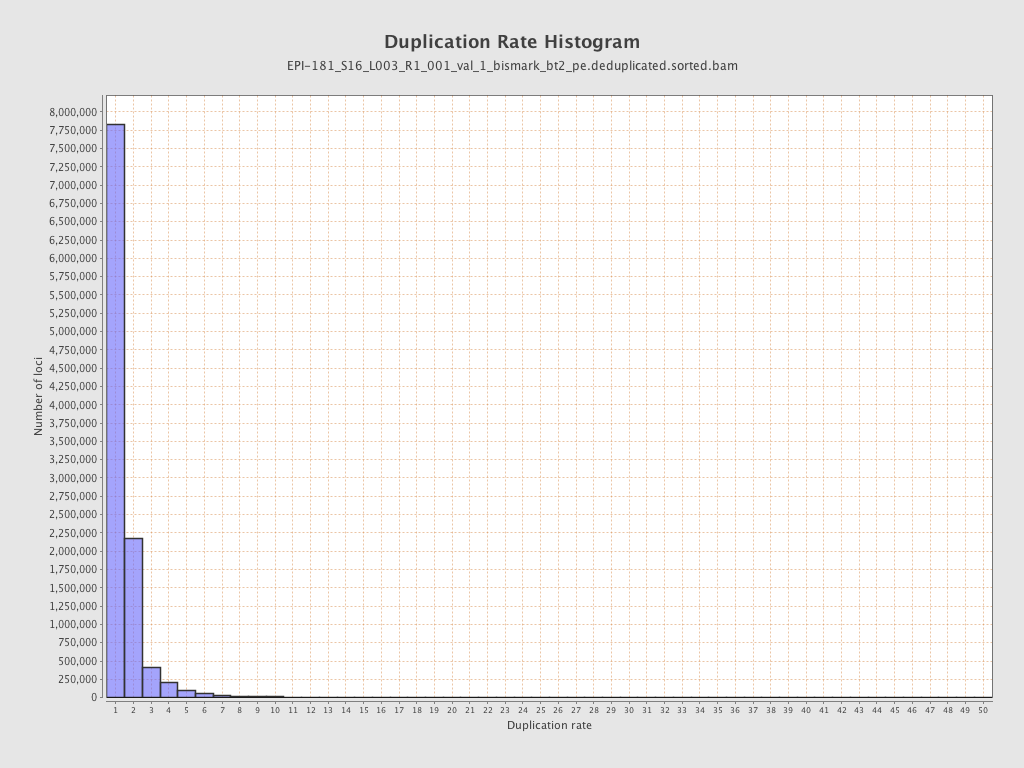
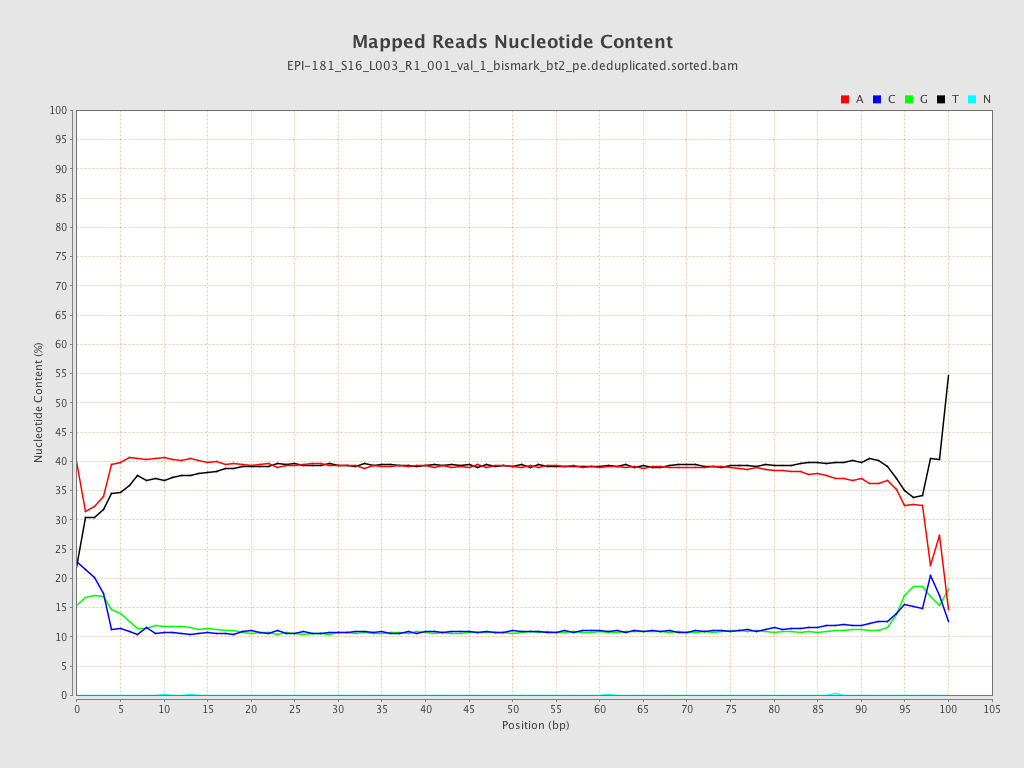
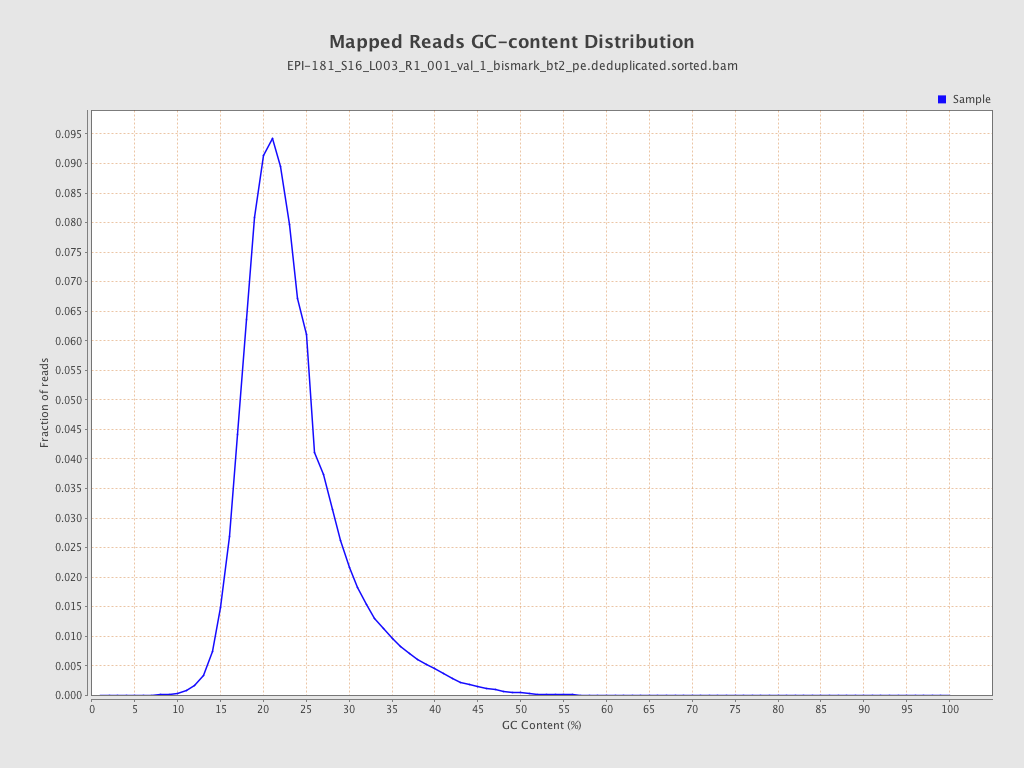
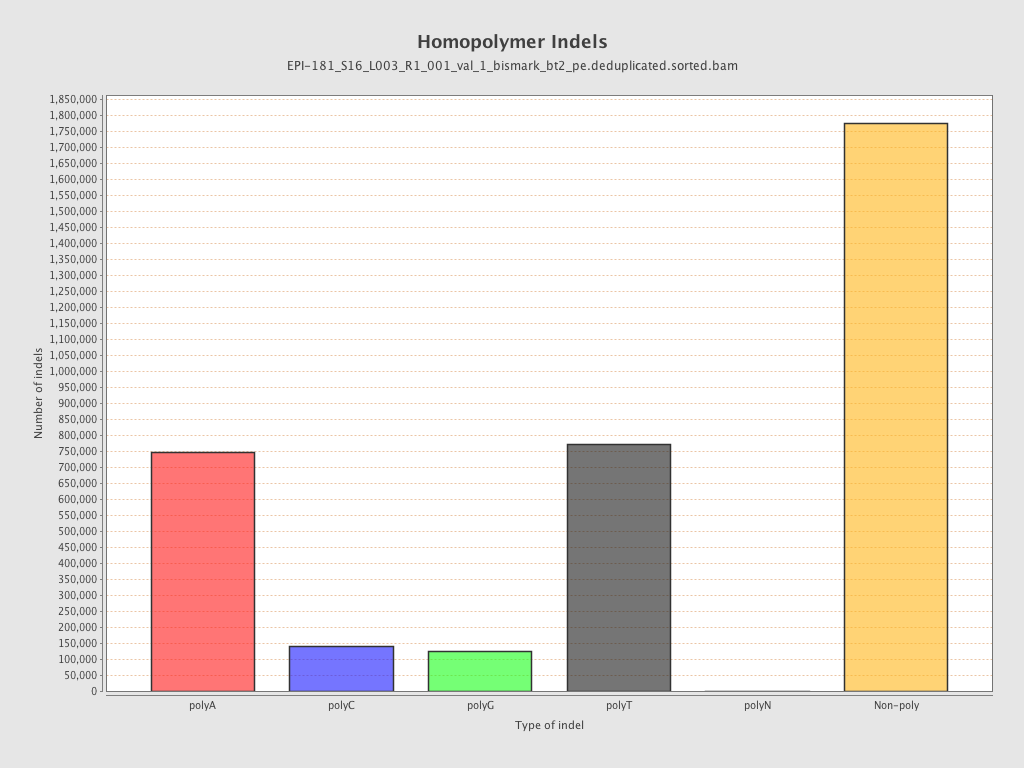
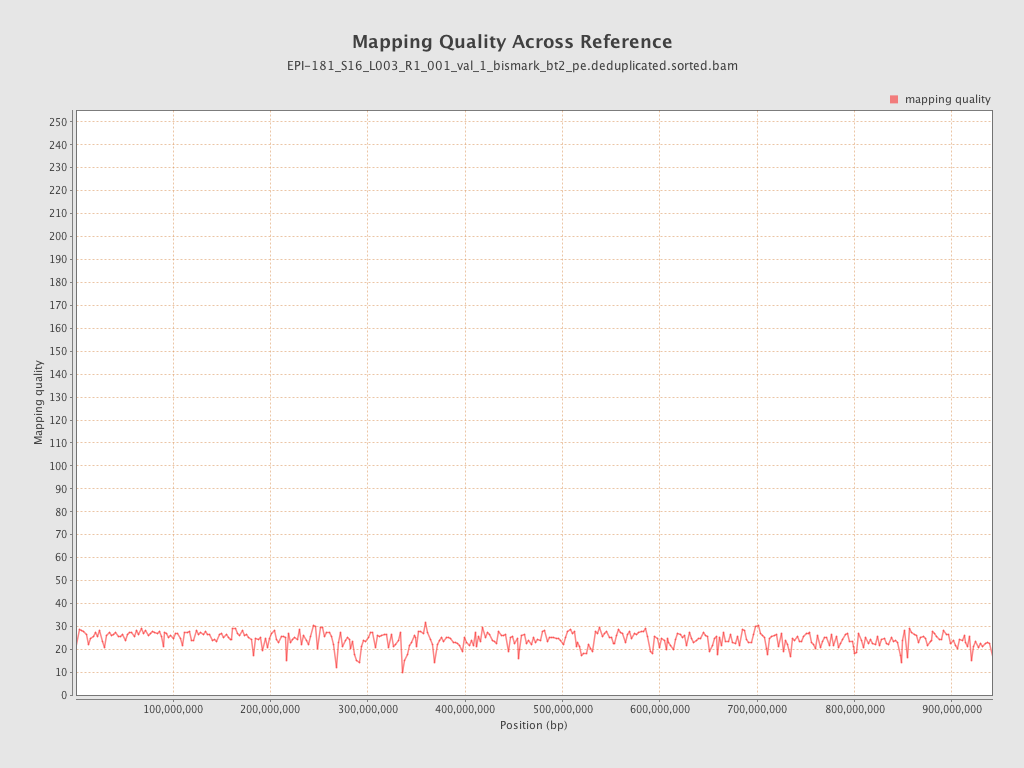
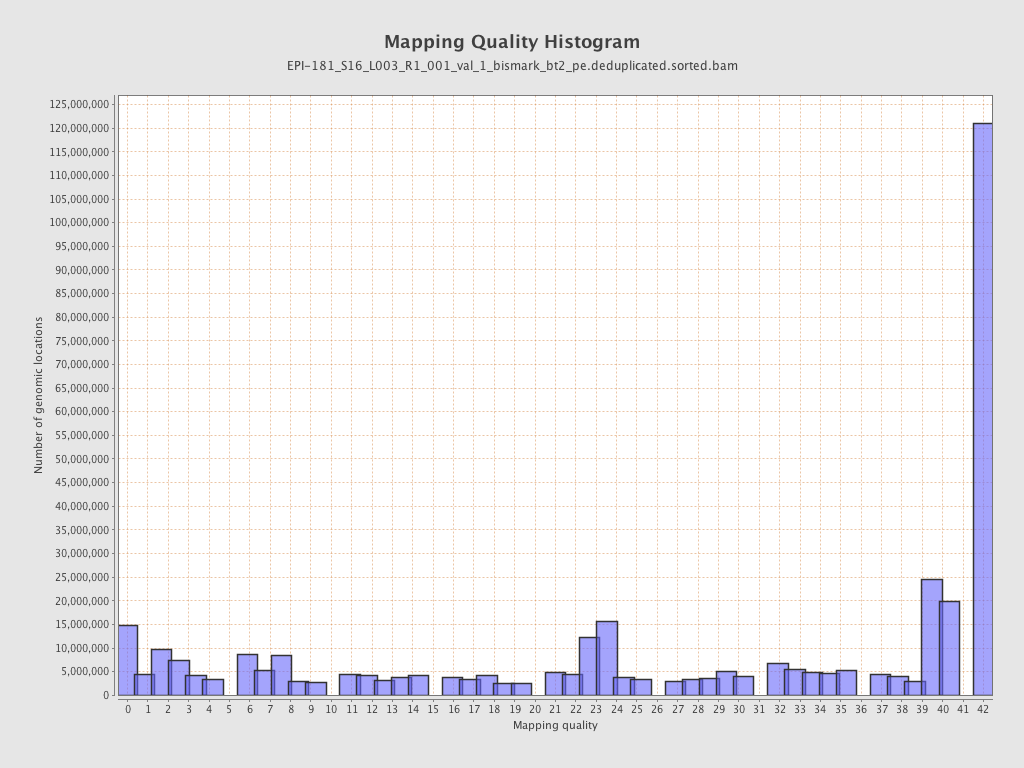
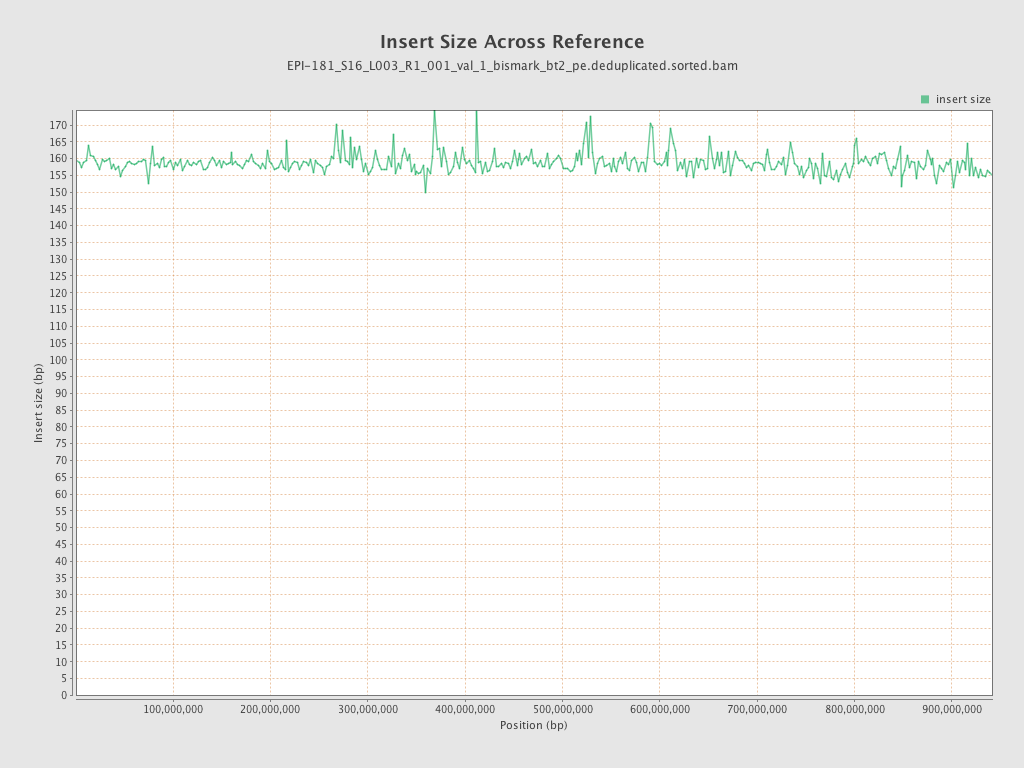
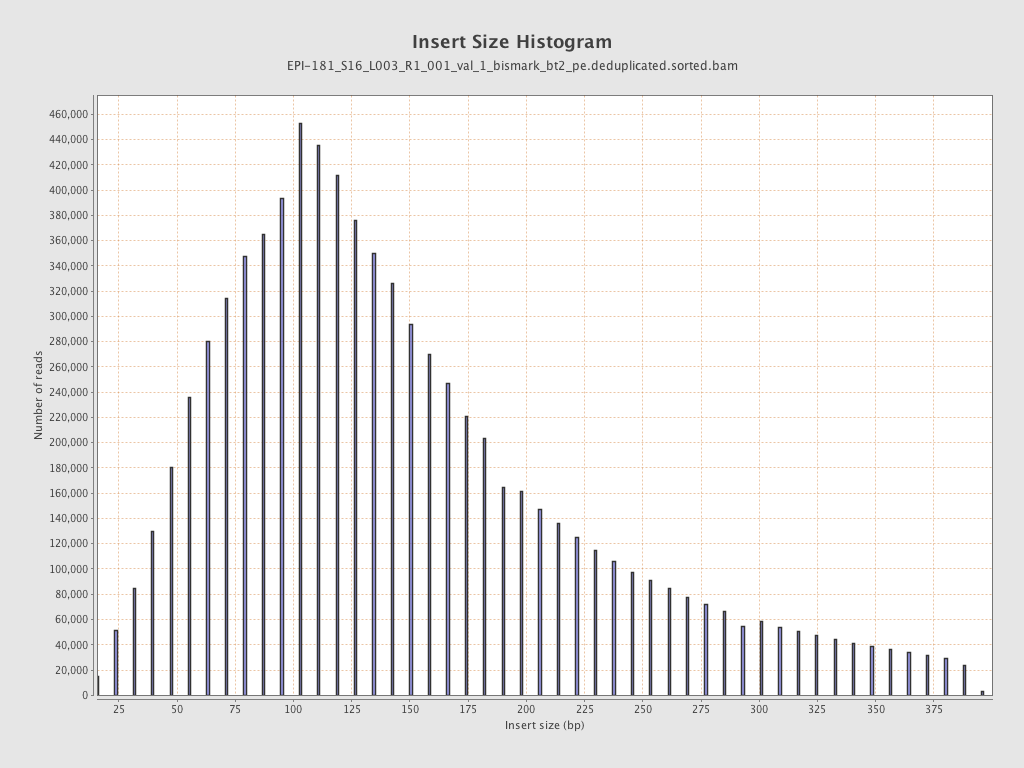
| Name |
Length |
Mapped bases |
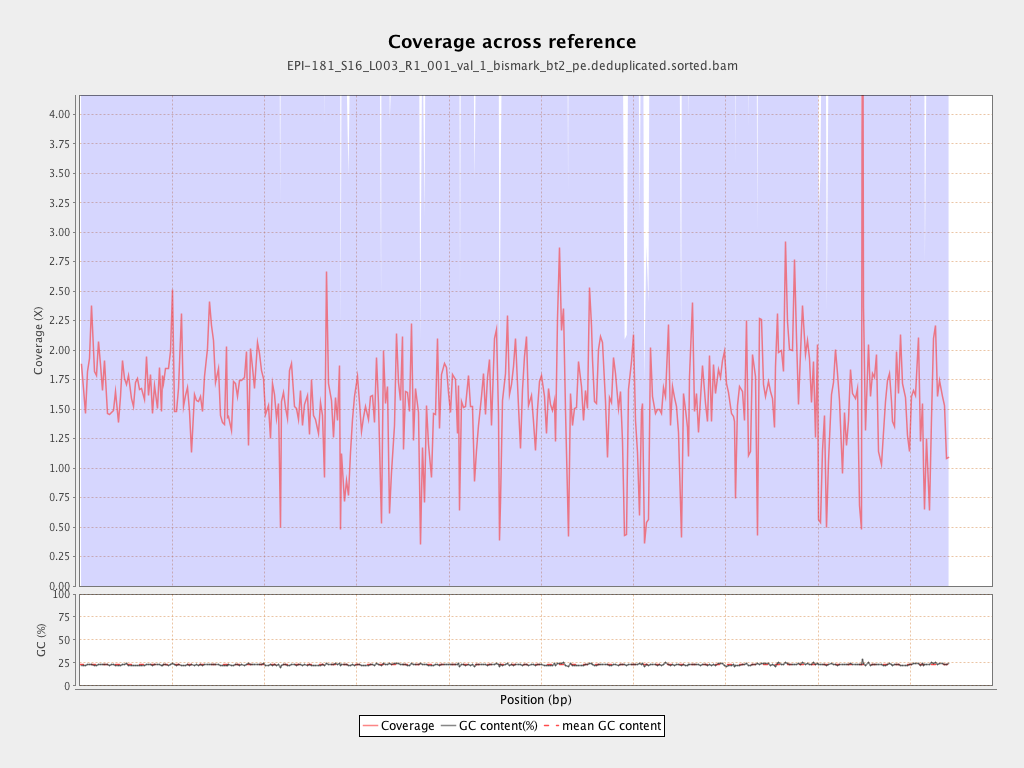
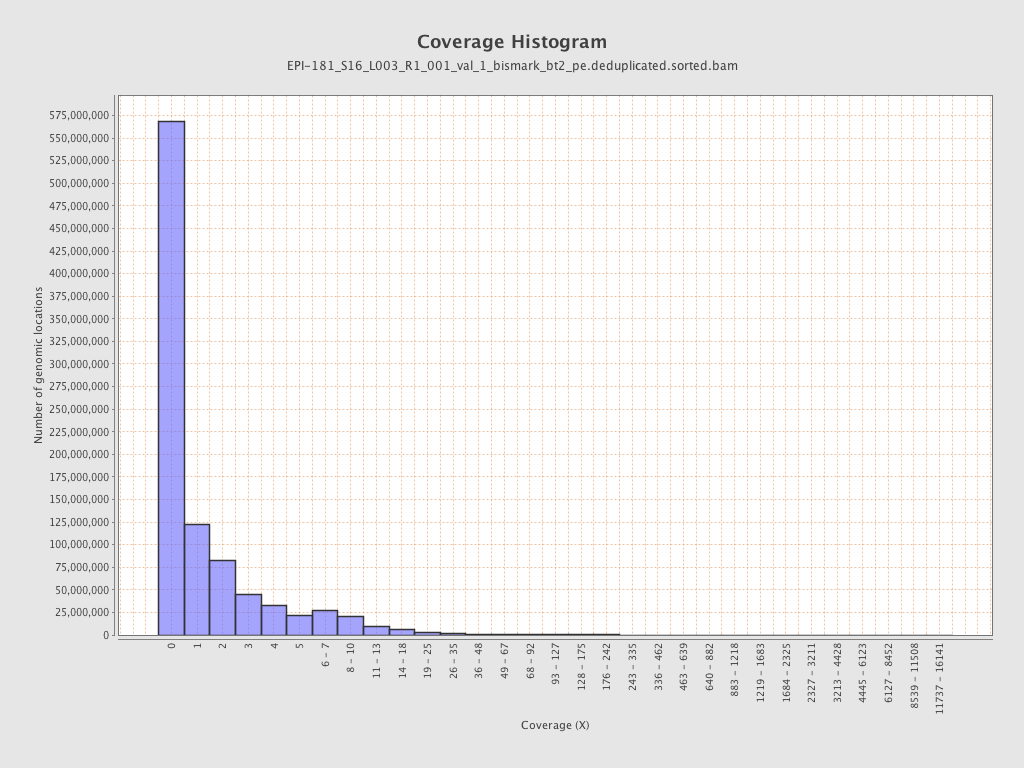
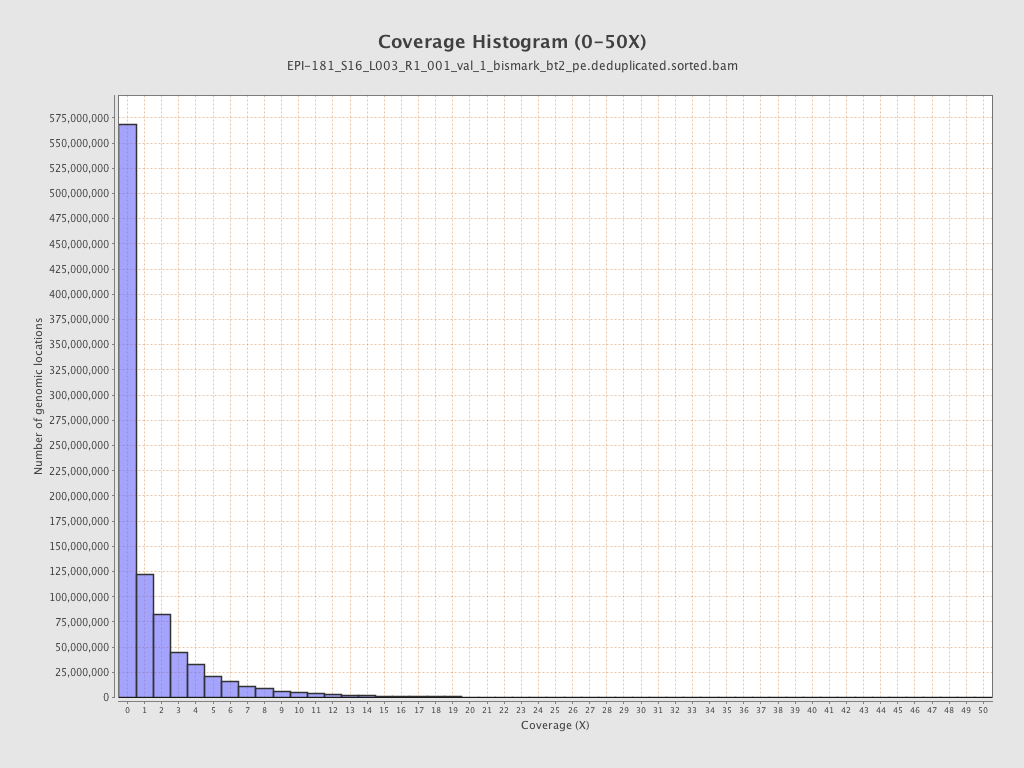
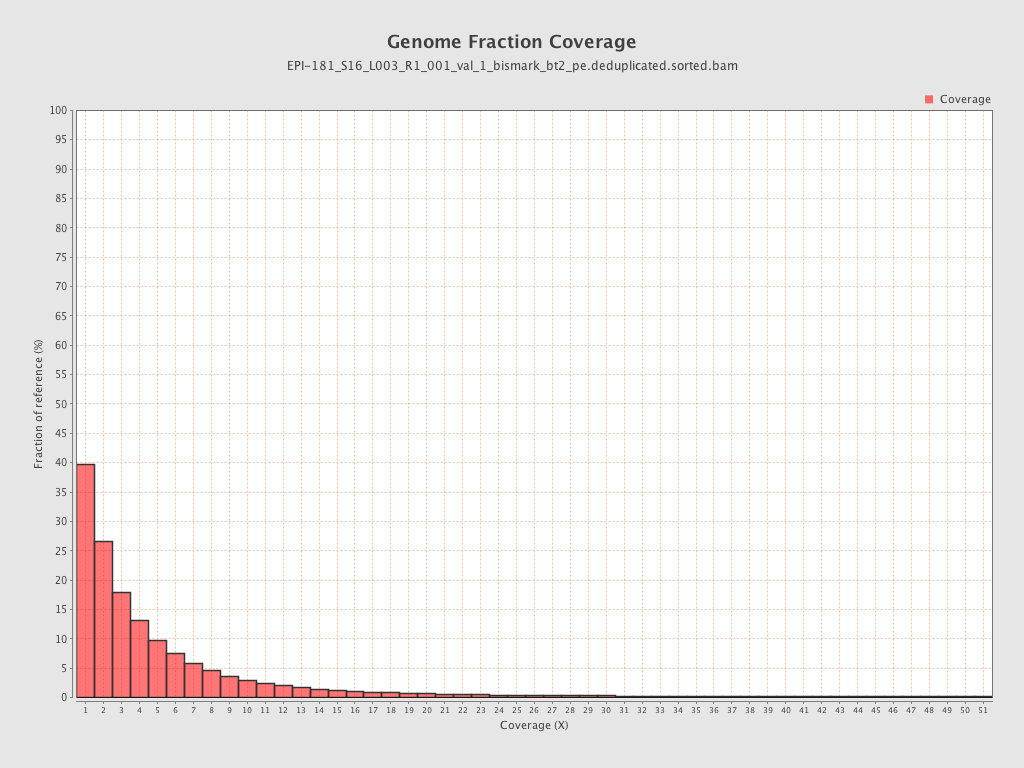
Mean coverage |
Standard deviation |
| PGA_scaffold1__77_contigs__length_89643857 |
89643857 |
152871014 |
1.7053 |
5.2476 |
| PGA_scaffold2__36_contigs__length_69596280 |
69596280 |
121320892 |
1.7432 |
6.8664 |
| PGA_scaffold3__111_contigs__length_57743597 |
57743597 |
95627995 |
1.6561 |
5.2738 |
| PGA_scaffold4__129_contigs__length_65288255 |
65288255 |
101681770 |
1.5574 |
7.5253 |
| PGA_scaffold5__109_contigs__length_67248332 |
67248332 |
93649126 |
1.3926 |
5.6851 |
| PGA_scaffold6__104_contigs__length_61759565 |
61759565 |
91403588 |
1.48 |
9.1166 |
| PGA_scaffold7__69_contigs__length_43120122 |
43120122 |
67265434 |
1.56 |
5.0117 |
| PGA_scaffold8__63_contigs__length_61151155 |
61151155 |
98355807 |
1.6084 |
5.7868 |
| PGA_scaffold9__45_contigs__length_38581958 |
38581958 |
68196474 |
1.7676 |
13.7836 |
| PGA_scaffold10__49_contigs__length_53961475 |
53961475 |
81570728 |
1.5116 |
4.6317 |
| PGA_scaffold11__79_contigs__length_51449921 |
51449921 |
71100407 |
1.3819 |
5.8454 |
| PGA_scaffold12__71_contigs__length_50438331 |
50438331 |
85306165 |
1.6913 |
6.8399 |
| PGA_scaffold13__52_contigs__length_44396874 |
44396874 |
70576509 |
1.5897 |
6.0799 |
| PGA_scaffold14__91_contigs__length_45393038 |
45393038 |
92384075 |
2.0352 |
25.8137 |
| PGA_scaffold15__101_contigs__length_47938513 |
47938513 |
62507910 |
1.3039 |
5.0769 |
| PGA_scaffold16__33_contigs__length_31980953 |
31980953 |
54722982 |
1.7111 |
18.2544 |
| PGA_scaffold17__51_contigs__length_34923512 |
34923512 |
55777543 |
1.5971 |
5.7678 |
| PGA_scaffold18__69_contigs__length_27737463 |
27737463 |
39549183 |
1.4258 |
8.0847 |