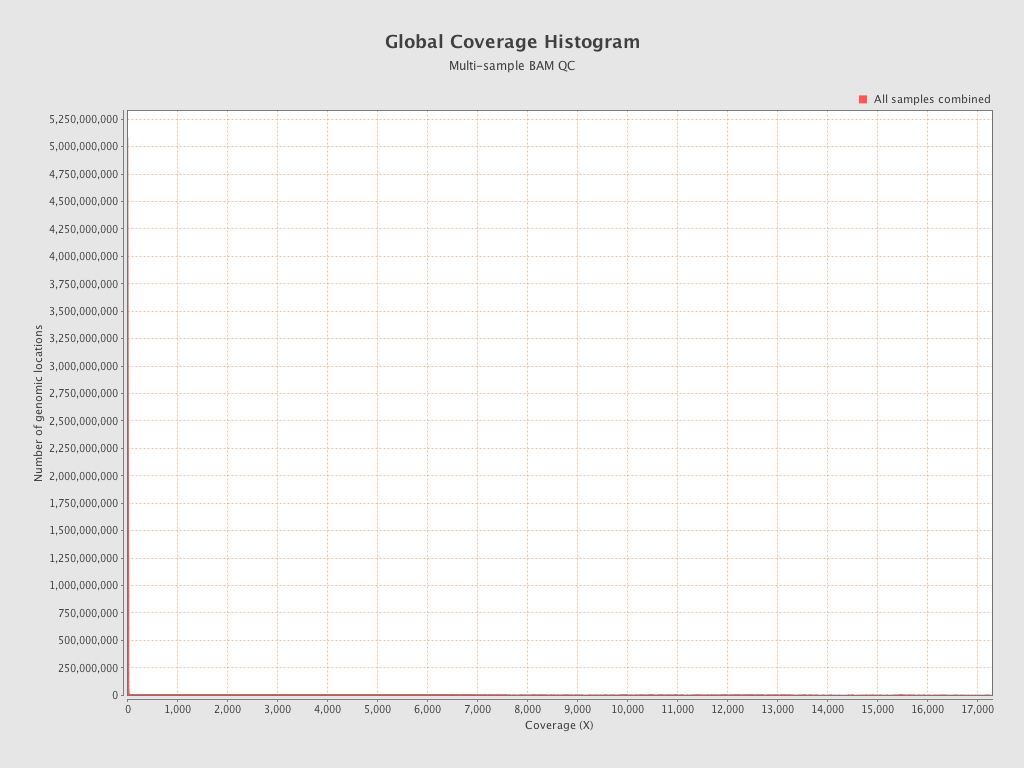
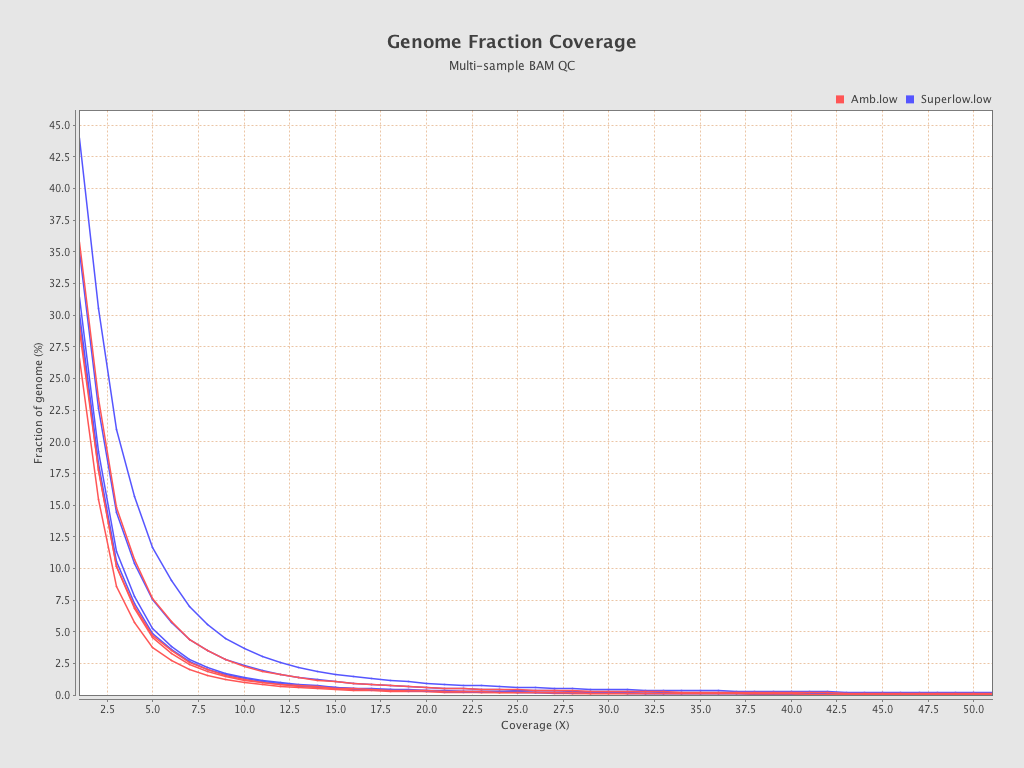
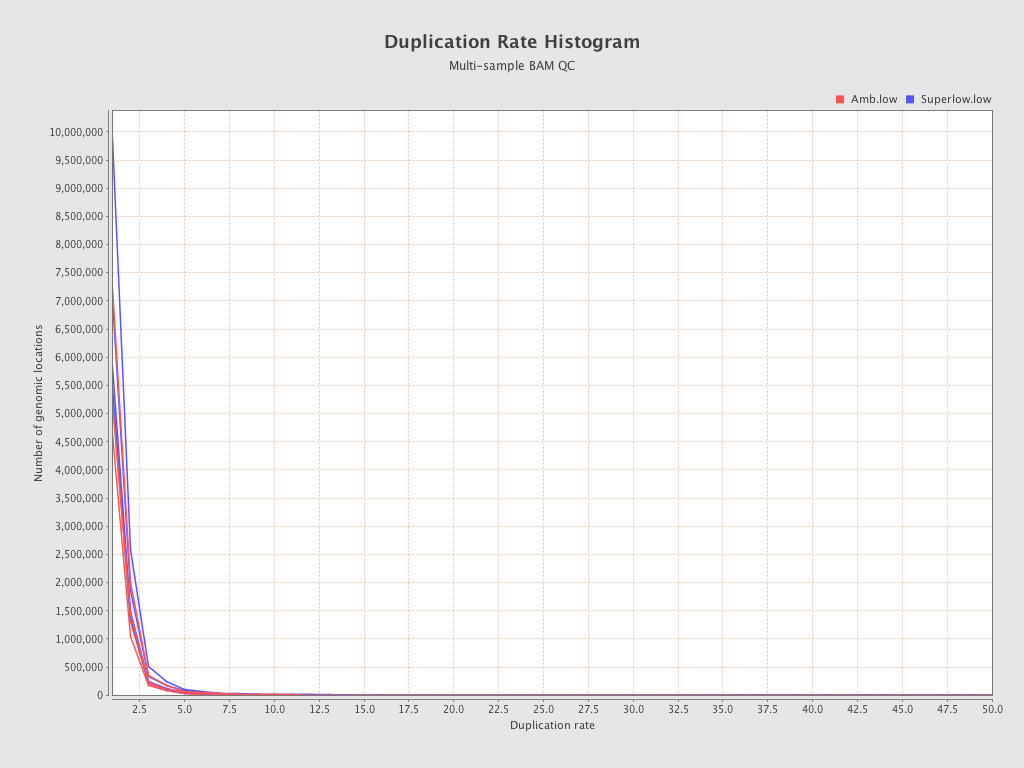
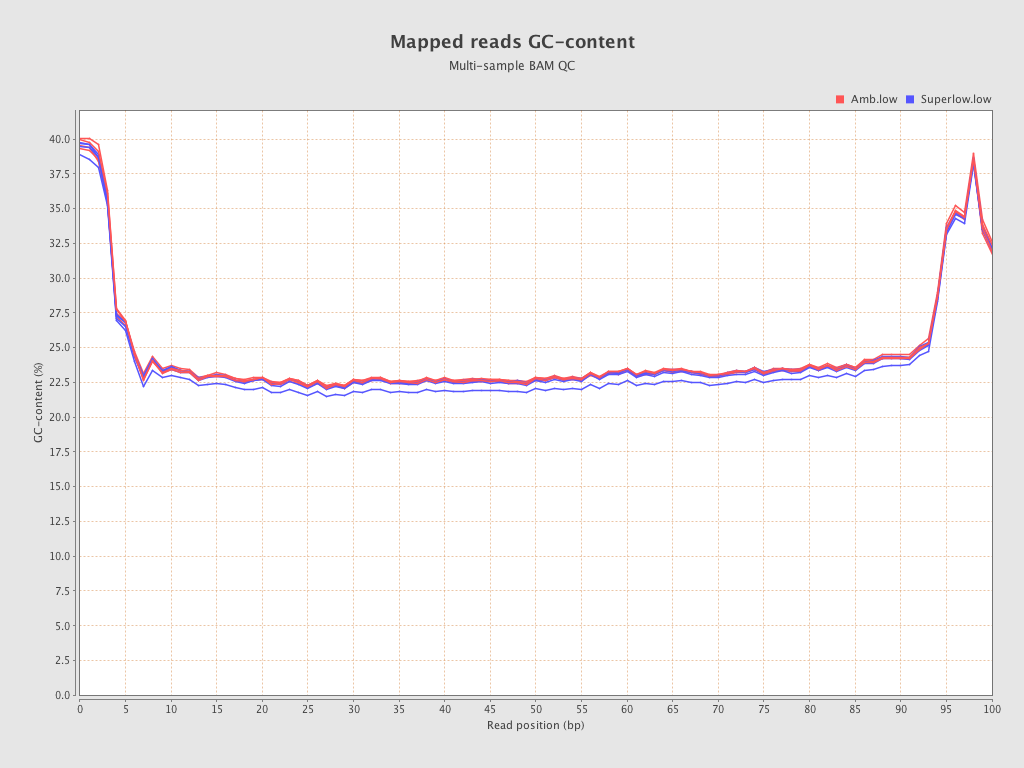
| 206 (Amb.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-206_S27_L004_dedup.sorted_stats |
| 205 (Amb.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-205_S26_L004_dedup.sorted_stats |
| 221 (Superlow.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-221_S33_L004_dedup.sorted_stats |
| 220 (Superlow.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-220_S32_L004_dedup.sorted_stats |
| 215 (Superlow.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-215_S31_L004_dedup.sorted_stats |
| 227 (Amb.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-227_S35_L004_dedup.sorted_stats |
| 226 (Amb.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-226_S34_L004_dedup.sorted_stats |
| 214 (Superlow.low) |
/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/EPI-214_S30_L004_dedup.sorted_stats |


.png)