06-cod-RNAseq-alignment
================
Kathleen Durkin
2024-03-19
- 1 Create a Bash variables
file
- 2
Align to reference transcriptome (Kallisto pseudoalignment)
- 2.1 Retrieving the
reference transcriptome
- 2.2 Verify
transcriptome FastA MD5 checksum
- 2.3 Building
Index
- 2.4 Sample Quantification
- 2.5 MultiQC on Kallisto
output logs
- 2.6 Trinity Matrix with
Kallisto Output
``` r
library(tidyverse)
library(dplyr)
library(magrittr)
library(knitr)
library(ggplot2)
library(plotly)
```
Code for aligning RNAseq data to reference transcriptome/genome, to be
used on [Pacific cod RNAseq
data](https://shedurkin.github.io/Roberts-LabNotebook/posts/projects/pacific_cod/2023_12_13_pacific_cod.html).
- trimmed reads generated in `05-cod-RNAseq-trimming`
- Transcriptome downloaded from
[NCBI](https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_031168955.1/),
stored
[here](https://owl.fish.washington.edu/halfshell/genomic-databank/GCF_031168955.1_ASM3116895v1_rna.fna)
as a part of lab [genomic
resources](https://robertslab.github.io/resources/Genomic-Resources/#gadus-macrocephalus-pacific-cod)
Note: Kallisto pseudoalignment doesn’t necessarily require input reads
to be trimmed, provided they are of sufficient quality.
# 1 Create a Bash variables file
This allows usage of Bash variables (e.g. paths to common directories)
across R Markdown chunks.
``` bash
{
echo "#### Assign Variables ####"
echo ""
echo "# Data directories"
echo 'export cod_dir=/home/shared/8TB_HDD_02/shedurkin/project-cod-temperature'
echo 'export output_dir_top=${cod_dir}/output/06-cod-RNAseq-alignment'
echo 'export trimmed_fastqc_dir=${cod_dir}/output/05-cod-RNAseq-trimming/trimmed-fastqc'
echo 'export trimmed_reads_dir=${cod_dir}/output/05-cod-RNAseq-trimming/trimmed-reads'
echo 'export kallisto_output_dir=${output_dir_top}/kallisto'
echo ""
echo "# Input/Output files"
echo 'export transcriptome_fasta_dir=${cod_dir}/data'
echo 'export transcriptome_fasta_name="GCF_031168955.1_ASM3116895v1_rna"'
echo 'export transcriptome_fasta="${transcriptome_fasta_dir}/${transcriptome_fasta_name}"'
echo 'export kallisto_index_name="G_macrocephalus_kallisto_index.idx"'
echo "# External data URLs and checksums"
echo 'export transcriptome_fasta_url="https://owl.fish.washington.edu/halfshell/genomic-databank/GCF_031168955.1_ASM3116895v1_rna.fna"'
echo 'export transcriptome_checksum="2a6c7c98982727e688f033a9b236725b"'
echo ""
echo "# Paths to programs"
echo 'export kallisto=/home/shared/kallisto/kallisto'
echo 'export multiqc=/home/sam/programs/mambaforge/bin/multiqc'
echo 'export trinity_abund_to_matrix=/home/shared/trinityrnaseq-v2.12.0/util/abundance_estimates_to_matrix.pl'
echo ""
echo "# Set number of CPUs to use"
echo 'export threads=20'
echo ""
echo "# Programs associative array"
echo "declare -A programs_array"
echo "programs_array=("
echo '[kallisto]="${kallisto}" \'
echo '[multiqc]="${multiqc}" \'
echo '[trinity_abund_to_matrix]="${trinity_abund_to_matrix}" \'
echo ")"
} > .bashvars
cat .bashvars
```
#### Assign Variables ####
# Data directories
export cod_dir=/home/shared/8TB_HDD_02/shedurkin/project-cod-temperature
export output_dir_top=${cod_dir}/output/06-cod-RNAseq-alignment
export trimmed_fastqc_dir=${cod_dir}/output/05-cod-RNAseq-trimming/trimmed-fastqc
export trimmed_reads_dir=${cod_dir}/output/05-cod-RNAseq-trimming/trimmed-reads
export kallisto_output_dir=${output_dir_top}/kallisto
# Input/Output files
export transcriptome_fasta_dir=${cod_dir}/data
export transcriptome_fasta_name="GCF_031168955.1_ASM3116895v1_rna"
export transcriptome_fasta="${transcriptome_fasta_dir}/${transcriptome_fasta_name}"
export kallisto_index_name="G_macrocephalus_kallisto_index.idx"
# External data URLs and checksums
export transcriptome_fasta_url="https://owl.fish.washington.edu/halfshell/genomic-databank/GCF_031168955.1_ASM3116895v1_rna.fna"
export transcriptome_checksum="2a6c7c98982727e688f033a9b236725b"
# Paths to programs
export kallisto=/home/shared/kallisto/kallisto
export multiqc=/home/sam/programs/mambaforge/bin/multiqc
export trinity_abund_to_matrix=/home/shared/trinityrnaseq-v2.12.0/util/abundance_estimates_to_matrix.pl
# Set number of CPUs to use
export threads=20
# Programs associative array
declare -A programs_array
programs_array=(
[kallisto]="${kallisto}" \
[multiqc]="${multiqc}" \
[trinity_abund_to_matrix]="${trinity_abund_to_matrix}" \
)
I will be running kallisto with trimmed/QCd reads
# 2 Align to reference transcriptome (Kallisto pseudoalignment)
## 2.1 Retrieving the reference transcriptome
``` bash
# Load bash variables into memory
source .bashvars
wget \
--directory-prefix ${transcriptome_fasta_dir} \
--recursive \
--no-check-certificate \
--continue \
--no-host-directories \
--no-directories \
--no-parent \
--quiet \
--execute robots=off \
--accept "${transcriptome_fasta_name}.fna" ${transcriptome_fasta_url}
```
``` bash
# Load bash variables into memory
source .bashvars
ls -lh "${transcriptome_fasta_dir}"
```
total 5.2G
drwxr-xr-x 3 shedurkin labmembers 4.0K Mar 4 11:05 05-cod-RNAseq-trimming
-rw-r--r-- 1 shedurkin labmembers 13K Dec 27 15:45 Cod_RNAseq_NGS_Template_File.xlsx
-rw-r--r-- 1 shedurkin labmembers 253 May 22 11:36 conditions.txt
-rw-r--r-- 1 shedurkin labmembers 2.1K Mar 20 20:55 DESeq2_Sample_Information.csv
-rw-r--r-- 1 shedurkin labmembers 38M Oct 25 2023 Gadus_macrocephalus.coding.gene.V1.cds
-rw-r--r-- 1 shedurkin labmembers 537M Oct 16 2023 GCF_031168955.1_ASM3116895v1_genomic.fna
-rw-r--r-- 1 shedurkin labmembers 875 May 7 14:05 GCF_031168955.1_ASM3116895v1_genomic.fna.fai
-rw-r--r-- 1 shedurkin labmembers 351M Oct 16 2023 GCF_031168955.1_ASM3116895v1.gff
-rw-r--r-- 1 shedurkin labmembers 169M Oct 16 2023 GCF_031168955.1_ASM3116895v1_rna.fna
-rw-r--r-- 1 shedurkin labmembers 404M Apr 23 14:29 genomic.gtf
-rw-r--r-- 1 shedurkin labmembers 1.5G May 8 15:44 Gmac_genes_fasta.fasta
-rw-r--r-- 1 shedurkin labmembers 1.5G May 8 16:14 Gmac_genes_fasta.tab
-rw-r--r-- 1 shedurkin labmembers 1.3K May 22 11:36 list01.txt
-rw-r--r-- 1 shedurkin labmembers 47K Oct 25 2023 Pcod Temp Growth experiment 2022-23 DATA.xlsx
-rw-r--r-- 1 shedurkin labmembers 1.6K May 22 11:36 README.md
-rw-r--r-- 1 shedurkin labmembers 231K Mar 4 17:41 Sample.QC.report.of_30-943133806_240118025106.pdf
-rw-r--r-- 1 shedurkin labmembers 12K Mar 4 17:41 Sample.QC.report.of_30-943133806_240118025106.xlsx
-rw-r--r-- 1 shedurkin labmembers 12K Oct 25 2023 temp-experiment.csv
-rw-r--r-- 1 shedurkin labmembers 271M Oct 25 2023 uniprot_sprot_r2023_04.fasta
-rw-r--r-- 1 shedurkin labmembers 88M May 8 10:57 uniprot_sprot_r2023_04.fasta.gz
-rw-r--r-- 1 shedurkin labmembers 415M May 8 10:57 uniprot_table_r2023_01.tab
## 2.2 Verify transcriptome FastA MD5 checksum
``` bash
# Load bash variables into memory
source .bashvars
cd "${transcriptome_fasta_dir}"
# Checksums file contains other files, so this just looks for the sRNAseq files.
md5sum --check <<< "${transcriptome_checksum} ${transcriptome_fasta_name}.fna"
```
GCF_031168955.1_ASM3116895v1_rna.fna: OK
## 2.3 Building Index
``` bash
# Load bash variables into memory
source .bashvars
cd "${kallisto_output_dir}"
${programs_array[kallisto]} index \
--threads=${threads} \
--index="${kallisto_index_name}" \
"${transcriptome_fasta}.fna"
```
``` bash
# Load bash variables into memory
source .bashvars
ls -lh ${kallisto_output_dir}
```
total 1.5G
-rw-r--r-- 1 shedurkin labmembers 1.5G Mar 18 16:08 G_macrocephalus_kallisto_index.idx
-rw-r--r-- 1 shedurkin labmembers 20M Jun 10 16:12 kallisto.isoform.counts.matrix
-rw-r--r-- 1 shedurkin labmembers 0 Jun 10 16:12 kallisto.isoform.TMM.EXPR.matrix
-rw-r--r-- 1 shedurkin labmembers 25M Jun 10 16:12 kallisto.isoform.TPM.not_cross_norm
-rw-r--r-- 1 shedurkin labmembers 532 Jun 10 16:12 kallisto.isoform.TPM.not_cross_norm.runTMM.R
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:50 kallisto_quant_1
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:43 kallisto_quant_10
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:34 kallisto_quant_100
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:35 kallisto_quant_100.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:36 kallisto_quant_107
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:36 kallisto_quant_107.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:38 kallisto_quant_108
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:39 kallisto_quant_108.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:41 kallisto_quant_109
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:41 kallisto_quant_109.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:43 kallisto_quant_10.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:54 kallisto_quant_11
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:45 kallisto_quant_110
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:45 kallisto_quant_110.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:47 kallisto_quant_117
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:48 kallisto_quant_117.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:49 kallisto_quant_118
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:50 kallisto_quant_118.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:52 kallisto_quant_119
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:52 kallisto_quant_119.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:55 kallisto_quant_11.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:06 kallisto_quant_12
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:56 kallisto_quant_120
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:57 kallisto_quant_120.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 12:59 kallisto_quant_121
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 12:59 kallisto_quant_121.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:01 kallisto_quant_127
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:02 kallisto_quant_127.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:03 kallisto_quant_128
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:03 kallisto_quant_128.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:04 kallisto_quant_129
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:04 kallisto_quant_129.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:07 kallisto_quant_12.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:15 kallisto_quant_13
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:08 kallisto_quant_131
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:08 kallisto_quant_131.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:09 kallisto_quant_137
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:09 kallisto_quant_137.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:11 kallisto_quant_138
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:11 kallisto_quant_138.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:12 kallisto_quant_139
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:13 kallisto_quant_139.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:15 kallisto_quant_13.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:16 kallisto_quant_140
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:17 kallisto_quant_140.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:18 kallisto_quant_147
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:18 kallisto_quant_147.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:20 kallisto_quant_148
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:21 kallisto_quant_148.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:29 kallisto_quant_149
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:31 kallisto_quant_149.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:34 kallisto_quant_150
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:34 kallisto_quant_150.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:36 kallisto_quant_18
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:37 kallisto_quant_18.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:39 kallisto_quant_19
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:42 kallisto_quant_19-G
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:43 kallisto_quant_19-G.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:40 kallisto_quant_19.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:46 kallisto_quant_19-S
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:47 kallisto_quant_19-S.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:51 kallisto_quant_1.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:11 kallisto_quant_2
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:53 kallisto_quant_20
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:56 kallisto_quant_20-G
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:57 kallisto_quant_20-G.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 13:53 kallisto_quant_20.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 13:59 kallisto_quant_20-S
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:00 kallisto_quant_20-S.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:02 kallisto_quant_21
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:03 kallisto_quant_21.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:05 kallisto_quant_28
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:06 kallisto_quant_28.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:08 kallisto_quant_29
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:09 kallisto_quant_29.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:12 kallisto_quant_2.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:28 kallisto_quant_3
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:14 kallisto_quant_30
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:15 kallisto_quant_30.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:17 kallisto_quant_31
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:18 kallisto_quant_31.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:20 kallisto_quant_37
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:20 kallisto_quant_37.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:22 kallisto_quant_38
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:23 kallisto_quant_38.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:25 kallisto_quant_39
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:26 kallisto_quant_39.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:29 kallisto_quant_3.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:45 kallisto_quant_4
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:31 kallisto_quant_40
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:32 kallisto_quant_40.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:33 kallisto_quant_41
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:34 kallisto_quant_41.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:36 kallisto_quant_47
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:37 kallisto_quant_47.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:39 kallisto_quant_48
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:39 kallisto_quant_48.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:42 kallisto_quant_49
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:43 kallisto_quant_49.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:46 kallisto_quant_4.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:13 kallisto_quant_5
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:48 kallisto_quant_50
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:49 kallisto_quant_50.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:51 kallisto_quant_57
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:55 kallisto_quant_57-G
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:56 kallisto_quant_57-G.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:51 kallisto_quant_57.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 14:57 kallisto_quant_57-S
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 14:58 kallisto_quant_57-S.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:00 kallisto_quant_58
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:04 kallisto_quant_58-G
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:06 kallisto_quant_58-G.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:01 kallisto_quant_58.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:07 kallisto_quant_58-S
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:08 kallisto_quant_58-S.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:10 kallisto_quant_59
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:11 kallisto_quant_59.log
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:14 kallisto_quant_5.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:16 kallisto_quant_60
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:17 kallisto_quant_60.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:19 kallisto_quant_67
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:20 kallisto_quant_67.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:22 kallisto_quant_68
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:23 kallisto_quant_68.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:25 kallisto_quant_69
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:26 kallisto_quant_69.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:27 kallisto_quant_70
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:28 kallisto_quant_70.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:30 kallisto_quant_78
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:31 kallisto_quant_78.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:33 kallisto_quant_79
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:33 kallisto_quant_79.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:35 kallisto_quant_80
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:36 kallisto_quant_80.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:38 kallisto_quant_83
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:38 kallisto_quant_83.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:40 kallisto_quant_88
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:41 kallisto_quant_88.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:43 kallisto_quant_90
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:43 kallisto_quant_90.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:46 kallisto_quant_91
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:46 kallisto_quant_91.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:48 kallisto_quant_92
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:48 kallisto_quant_92.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:50 kallisto_quant_97
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:51 kallisto_quant_97.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:53 kallisto_quant_98
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:54 kallisto_quant_98.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:55 kallisto_quant_99
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:56 kallisto_quant_99.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 15:58 kallisto_quant_RESUB-116
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 15:58 kallisto_quant_RESUB-116.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 16:00 kallisto_quant_RESUB-156
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 16:01 kallisto_quant_RESUB-156.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 16:02 kallisto_quant_RESUB-36
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 16:03 kallisto_quant_RESUB-36.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 16:04 kallisto_quant_RESUB-76
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 16:05 kallisto_quant_RESUB-76.log
drwxr-xr-x 2 shedurkin labmembers 4.0K Jun 10 16:07 kallisto_quant_RESUB-94
-rw-r--r-- 1 shedurkin labmembers 5.3K Jun 10 16:07 kallisto_quant_RESUB-94.log
## 2.4 Sample Quantification
Kallisto can run quantification on either single- or paired-end reads.
The default option is paired-end, which requires the input of an even
number of paired fastq files (e.g., pairA_R1.fastq, pairA_R2.fastq). To
use single-end mode, include the –single flag, as well as -l
(–fragment-length=DOUBLE, estimated avg. fragment length) and -s
(–sd=DOUBLE, estimates stand. dev. of fragment length), and a number of
fastq files. Again, gzipped files are acceptable.
Kallisto quant is rather finicky about how you input sets of paired
reads, and you can only input a single pair at a time. To circumvent,
I’ll create a quantification function and apply it iteratively to each
pair of reads using a loop.
``` bash
# Load bash variables into memory
source .bashvars
# Function to run kallisto quant. Takes two (paired) reads as input, outputs to sample-associated directory
run_kallisto_quant() {
source .bashvars # Source .bashvars inside the function to make its variables accessible
local R1_fastq=${1}
local R2_fastq=${2}
cd ${kallisto_output_dir}
sample_num=$(basename "${R1_fastq}" ".flexbar_trim.R_1.fastq.gz")
mkdir kallisto_quant_${sample_num}
${programs_array[kallisto]} quant \
--threads=${threads} \
--index="${kallisto_output_dir}/${kallisto_index_name}" \
--output-dir="${kallisto_output_dir}/kallisto_quant_${sample_num}" \
--bootstrap-samples=100 \
${trimmed_reads_dir}/${R1_fastq} ${trimmed_reads_dir}/${R2_fastq} \
&> "${kallisto_output_dir}/kallisto_quant_${sample_num}.log"
}
# Iteratively apply run_kallisto_quant on each pair of input reads
for file_r1 in "${trimmed_reads_dir}"/*.flexbar_trim.R_1.fastq.gz; do
# Extract the sample name from the file name
sample_name=$(basename "${file_r1}" ".flexbar_trim.R_1.fastq.gz")
# Form the file names (function takes input file names, not paths)
file_r1_name="${sample_name}.flexbar_trim.R_1.fastq.gz"
file_r2_name="${sample_name}.flexbar_trim.R_2.fastq.gz"
# Check that the sample hasn't already been quantified
if [ ! -d "${kallisto_output_dir}/kallisto_quant_${sample_name}" ]; then
# Check if the corresponding R2 file exists
if [ -e "${trimmed_reads_dir}/${file_r2}" ]; then
# Run kallisto quant on the file pair
run_kallisto_quant "${file_r1_name}" "${file_r2_name}"
echo "Processed sample: ${sample_name}"
fi
else
echo "Sample already processed: ${sample_name}"
fi
done
```
Check that we have the appropriate number of output folders. We should
have one log file for each pair of reads
``` bash
# Load bash variables into memory
source .bashvars
# Count number of raw read files
cd ${trimmed_reads_dir}
echo "Number of raw reads:"
ls -1 *.fastq.gz | wc -l
# Count number of kallisto output
cd ${kallisto_output_dir}
echo "Number of output log files"
find . -type f -name "*.log" | wc -l
```
Number of raw reads:
160
Number of output log files
80
## 2.5 MultiQC on Kallisto output logs
``` bash
# Load bash variables into memory
source .bashvars
############ RUN MULTIQC ############
echo "Beginning MultiQC on raw FastQC..."
echo ""
${programs_array[multiqc]} ${kallisto_output_dir}/*.log -o ${output_dir_top}
echo ""
echo "MultiQC on raw FastQs complete."
echo ""
############ END MULTIQC ############
echo "Removing FastQC zip files."
echo ""
rm ${output_dir_top}/*.zip
echo "FastQC zip files removed."
echo ""
# View directory contents
ls -lh ${output_dir_top}
```
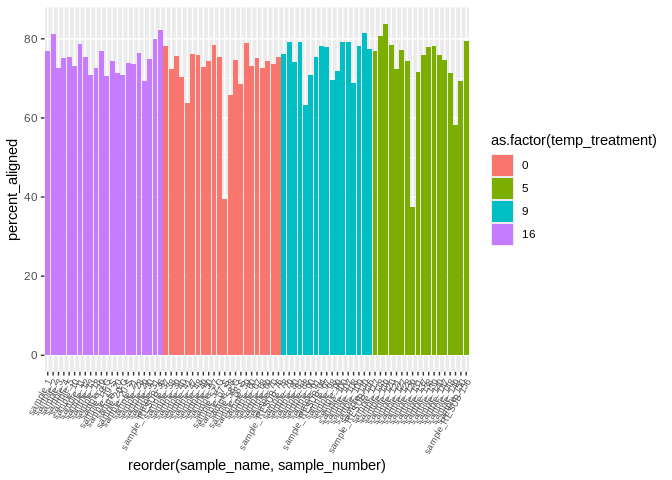
I also want to include the treatment/tank info when plotting alignment
rates across samples
``` r
# Load multiqc stats
kallisto_multiqc <- read.csv("../output/06-cod-RNAseq-alignment/multiqc_data/multiqc_kallisto.txt", sep = '\t')
# Adjust sample name formatting (to prep for join)
kallisto_multiqc$Sample <- gsub("_R1_001", "", kallisto_multiqc$Sample)
kallisto_multiqc$Sample <- paste("sample_", kallisto_multiqc$Sample, sep = "")
# Load experimental data
cod_sample_info_OG <- read.csv("../data/DESeq2_Sample_Information.csv")
kallisto_multiqc_plustreatment <- left_join(cod_sample_info_OG, kallisto_multiqc, by = c("sample_name" = "Sample")) %>%
na.omit()
kallisto_multiqc_plustreatment <- kallisto_multiqc_plustreatment[order(kallisto_multiqc_plustreatment$sample_number),]
ggplot(kallisto_multiqc_plustreatment,
aes(x=reorder(sample_name, sample_number), y=percent_aligned, fill=as.factor(temp_treatment))) +
geom_bar(stat="identity") +
theme(axis.text.x = element_text(angle = 60, vjust = 1, hjust=1, size=7))
```
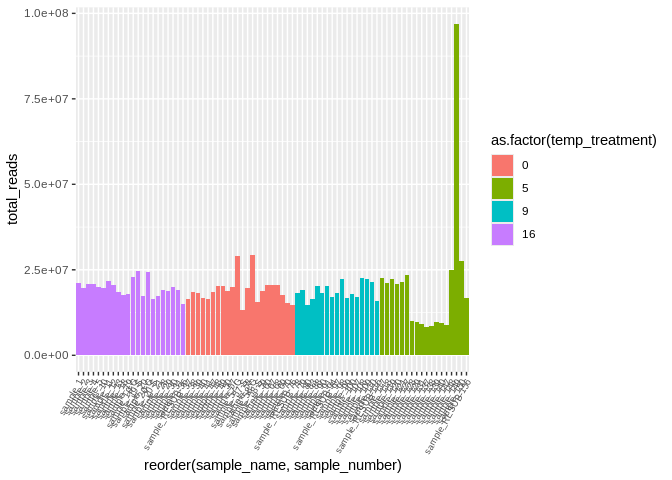
``` r
ggplot(kallisto_multiqc_plustreatment,
aes(x=reorder(sample_name, sample_number), y=total_reads, fill=as.factor(temp_treatment))) +
geom_bar(stat="identity") +
theme(axis.text.x = element_text(angle = 60, vjust = 1, hjust=1, size=7))
```
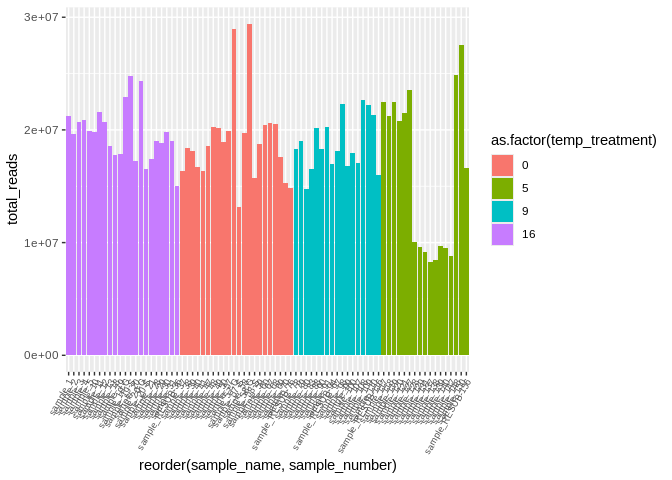
``` r
# sample149 is kind of throwing off the visualization, so lets remove and redo
ggplot(kallisto_multiqc_plustreatment[kallisto_multiqc_plustreatment$sample_name != "sample_149", ],
aes(x=reorder(sample_name, sample_number), y=total_reads, fill=as.factor(temp_treatment))) +
geom_bar(stat="identity") +
theme(axis.text.x = element_text(angle = 60, vjust = 1, hjust=1, size=7))
```
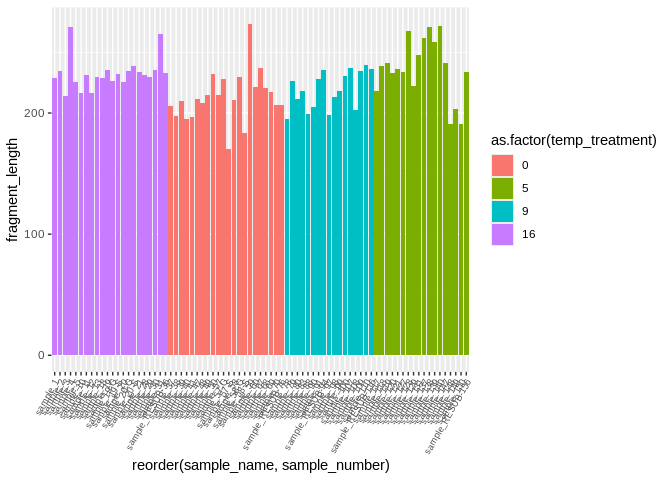
``` r
ggplot(kallisto_multiqc_plustreatment,
aes(x=reorder(sample_name, sample_number), y=fragment_length, fill=as.factor(temp_treatment))) +
geom_bar(stat="identity") +
theme(axis.text.x = element_text(angle = 60, vjust = 1, hjust=1, size=7))
```
## 2.6 Trinity Matrix with Kallisto Output
``` bash
# Load bash variables into memory
source .bashvars
cd ${kallisto_output_dir}
${programs_array[trinity_abund_to_matrix]} \
--est_method 'kallisto' \
--gene_trans_map 'none' \
--out_prefix 'kallisto' \
--name_sample_by_basedir ${kallisto_output_dir}/kallisto_quant_*/abundance.tsv
ls -lh ${kallisto_output_dir}
```