08-week8
#Week 08 Questions
What is a genomic range and what 3 types of information do you need for a range?
Genomic range describes the location, or coordinates on a genome of a given feature. A range is defined by the scaffold of the genome on which it’s found (e.g., the chromosome), the start position (e.g., # of nucleotides from the beginning of the scaffold to the beginning of the feature), and the end position (e.g., # nucleotides from beginning of scaffold to the end of the feature).
What does 0-based and 1-based refer to? What are the advantages and disadvantages of each?
0-based and 1-based refer to the two common systems of numbering base positions in genomic coordinate systems. In 0-based, the first base position is indexed as 0, in 1-based the first position is indexed as 1. Neither is particularly more or less advantageous, it’s just a distinction to be wary of.
What is the value of BEDtools over the bioconductor package GenomicRange?
BEDtools and GenomicRange provide very similar tools, but BEDtools is implemented through the command line, which can improve its computational flexibility.
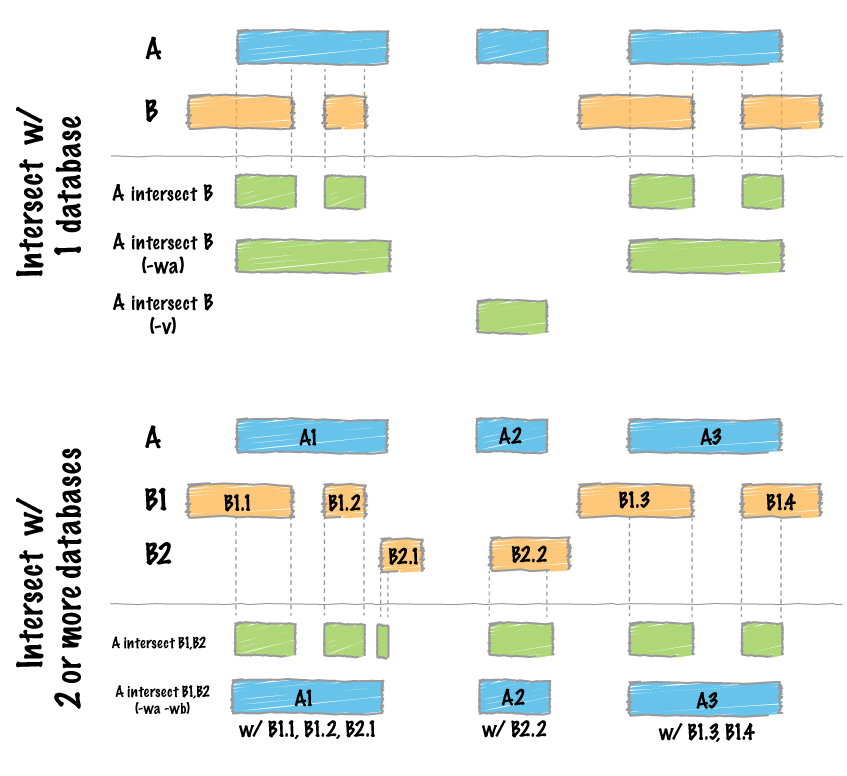
Describe one subcommand of the BEDtools suite as well as a practical use case.
intersect is a command that, given two input BED files (sets of genomic features), will identify overlapping regions that are present in both sets. For example, I’ve used this in my class project to find larval DMLs that were also present in the parental methylation landscapes, by finding the intersections of a BED file of my larval DMLs and a BED file of parental DMLs.