Input data and parameters
Input
| Analysis date: | Fri Mar 25 20:37:36 GMT 2022 |
| BAM file: | ctenidia.markdup.sorted.bam |
| Counting algorithm: | uniquely-mapped-reads |
| GTF file: | Panopea-generosa-v1.0.a4_biotype-trna_strand_converted-no_RNAmmer.gtf |
| Number of bases for 5'-3' bias computation: | 100 |
| Number of transcripts for 5'-3' bias computation: | 1,000 |
| Paired-end sequencing: | yes |
| Protocol: | strand-specific-reverse |
| Sorting performed: | yes |
Summary
Reads alignment
| Number of mapped reads (left/right): | 36,725,984 / 36,728,439 |
| Number of aligned pairs (without duplicates): | 36,685,637 |
| Total number of alignments: | 80,553,129 |
| Number of secondary alignments: | 7,098,706 |
| Number of non-unique alignments: | 11,599,239 |
| Aligned to genes: | 16,806,775 |
| Ambiguous alignments: | 1,066 |
| No feature assigned: | 52,146,049 |
| Not aligned: | 0 |
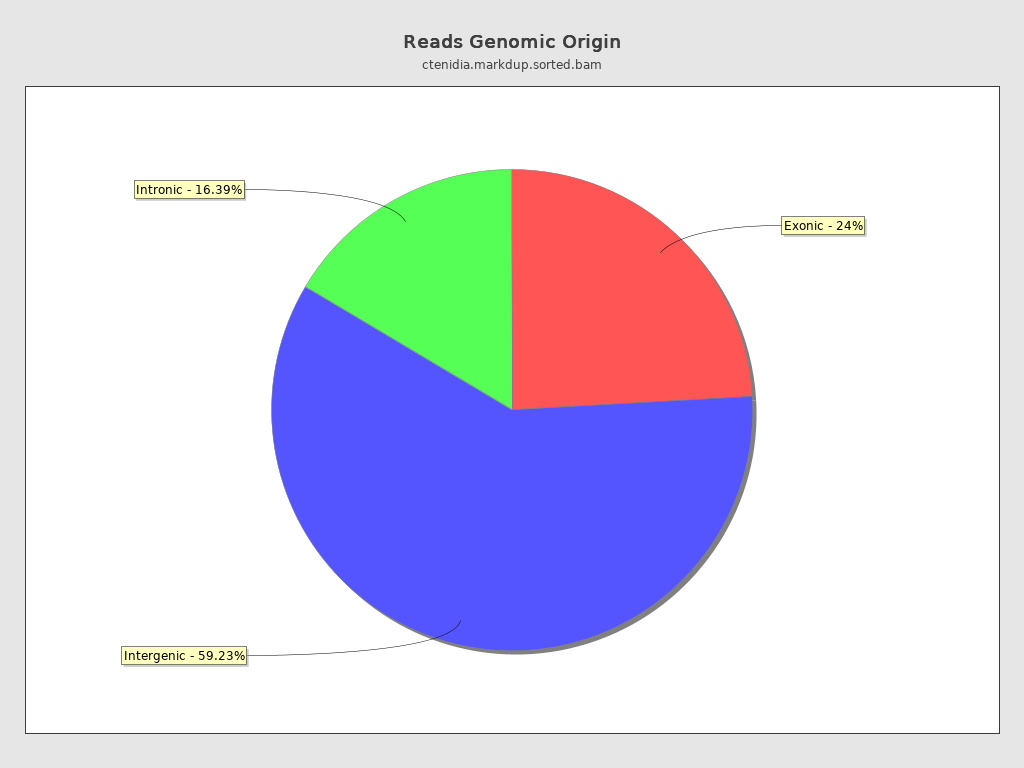
Reads genomic origin
| Exonic: | 16,806,775 / 24.37% |
| Intronic: | 11,301,982 / 16.39% |
| Intergenic: | 40,844,067 / 59.23% |
| Intronic/intergenic overlapping exon: | 4,132,563 / 5.99% |
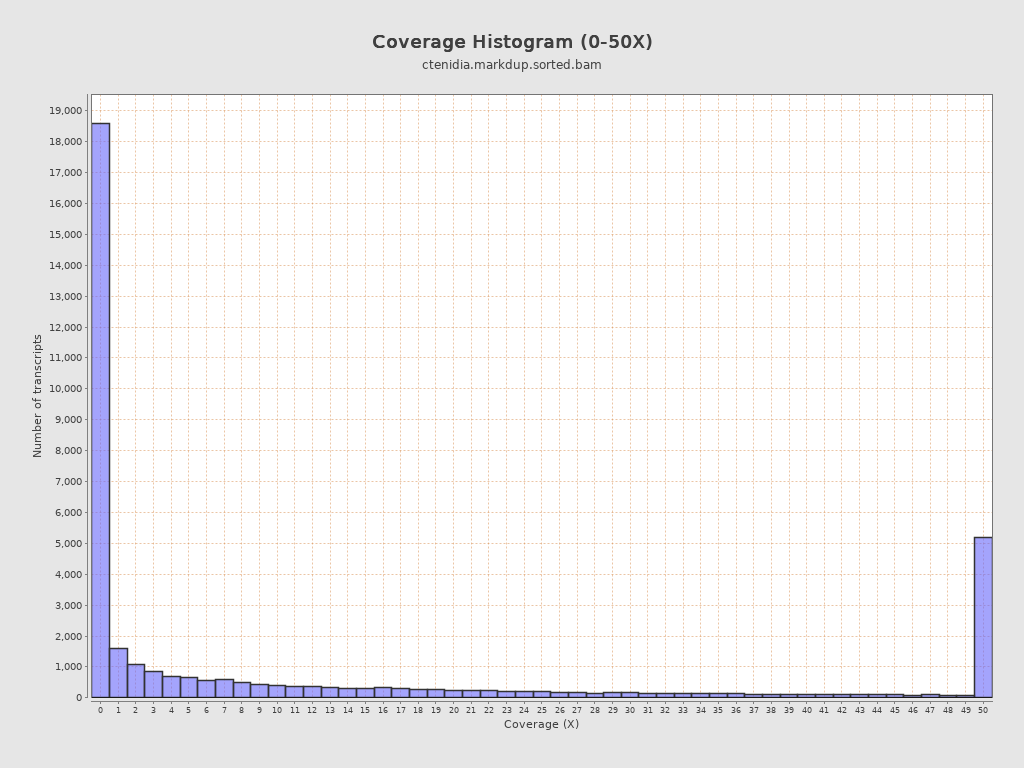
Transcript coverage profile
| 5' bias: | 0.52 |
| 3' bias: | 0.54 |
| 5'-3' bias: | 0.97 |
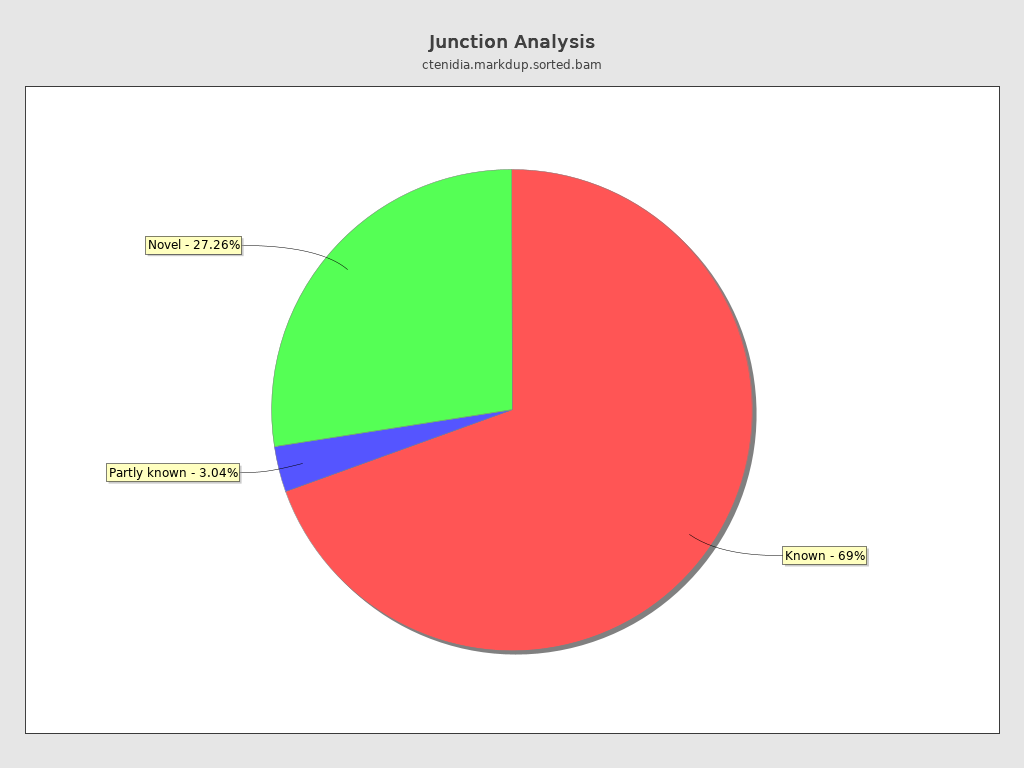
Junction analysis
| Reads at junctions: | 17,270,876 |
| AGGT | 6.9% |
| ACCT | 6.28% |
| TCCT | 3.44% |
| AGAT | 3.02% |
| ATCT | 2.89% |
| AGCT | 2.63% |
| AGGA | 2.53% |
| AGAA | 2.19% |
| AGGG | 2.08% |
| TTCT | 1.72% |
| AGGC | 1.66% |

.png)
.png)
.png)